Hung-Ta Chen Lab

Contact
- htchen@imb.sinica.edu.tw
- (L) 886-2-2789-9327
- N311, Institute of Molecular Biology, Academia Sinica
Research
Eukaryotic RNA Polymerase III Transcription
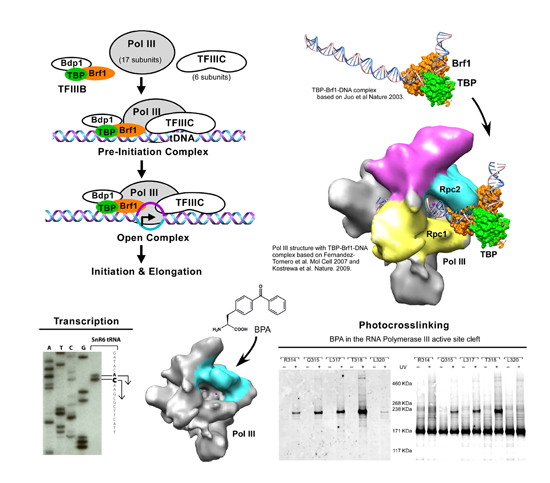
The research focus of our group is to understand the mechanism of eukaryotic RNA polymerase III (Pol III) transcription. Pol III is responsible for the transcription of tRNAs, 5S ribosomal RNA, and U6 spliceosomal RNA. Pol III relies on transcription factors TFIIIA, TFIIIB, and TFIIIC for the formation of the initiation complex at the gene promoter. We apply site-specific photocrosslinking and hydroxyl-radical probing analyses to map the binding targets for individual proteins in the initiation complex. The in vitro biochemical probing is conducted in the initiation complex assembled with proteins from the model organism Saccharomyces cerevisiae (yeast). We have successfully applied the mapping analyses to Pol III and TFIIIB with results allowing us to obtain the domain architecture of the initiation complex and explain how individual protein domains function in Pol III transcription.
Copyright © 2017 IMBCC. All rights reserved. |