
Contact
- jleu@imb.sinica.edu.tw
- (O) 886-2-2651-9574
(L) 886-2-2789-9216 - N411, Institute of Molecular Biology, Academia Sinica
Research
General principles and molecular mechanisms of endosymbiosis, phenotypic robustness, and speciation
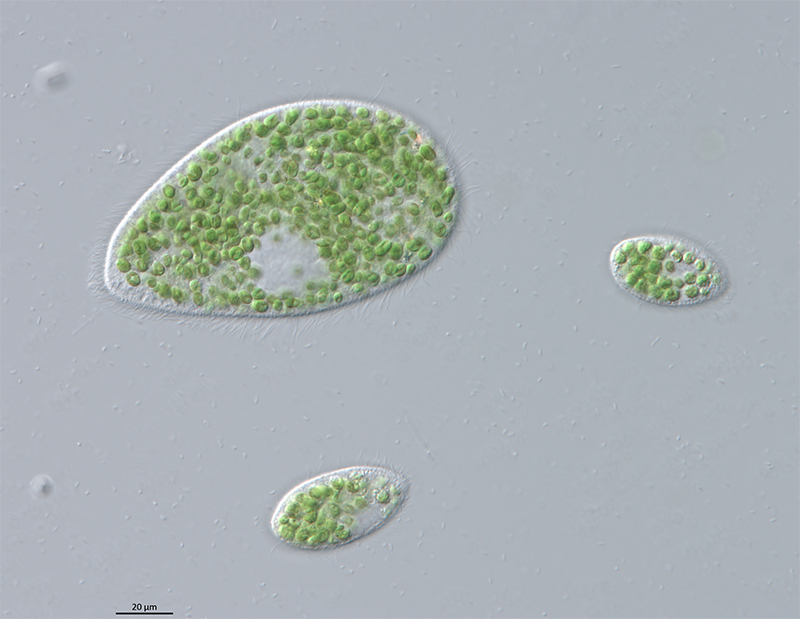
I. Genomic analysis of endosymbiosis between Paramecium and Chlorella
Why can an organism live inside another organism without causing trouble?
Endosymbiosis is one of the major forces driving the evolution of eukaryotic cells, which has occurred multiple times in different lineages during evolutionary history. However, the initial process of how the endosymbiosis is formed and established in the host cell is still unclear.
We use the ciliates Paramecium bursaria and Tetrahymena utriculariae, and the algae Chlorella spp. to study the early stage of endosymbiosis. Ciliates are an ancient and diverse group of protists exhibiting extraordinary features of cell biology. The green algae Chlorella spp. have been utilized in many industrial applications such as biofuel production. These two systems allow us to explore the genetic basis of endosymbiosis establishment and also other aspects of biology.
II. Molecular mechanisms and the evolution of phenotypic robustness
Why can the cell maintain consistent performance under a variety of conditions and genetic backgrounds?
The ability to maintain a stable phenotype despite environmental and genetic perturbations is a fundamental property of living systems. It has been speculated that cells contain master buffering systems that can establish phenotypic robustness under general conditions and regulate it under stress. Buffering systems are crucial in stabilizing cell physiology and developmental processes from environmental and genetic perturbations. Besides, buffering systems may play an important role in setting the tempo of evolution since they can moderate these perturbations or help a population accumulate genetic variations essential for selection.
Although the concept of genetic and non-genetic buffering is well established, the underlying mechanisms are largely unclear. In our lab, a systematic approach combining experimental evolution and genomic analysis is used to understand their molecular basis.
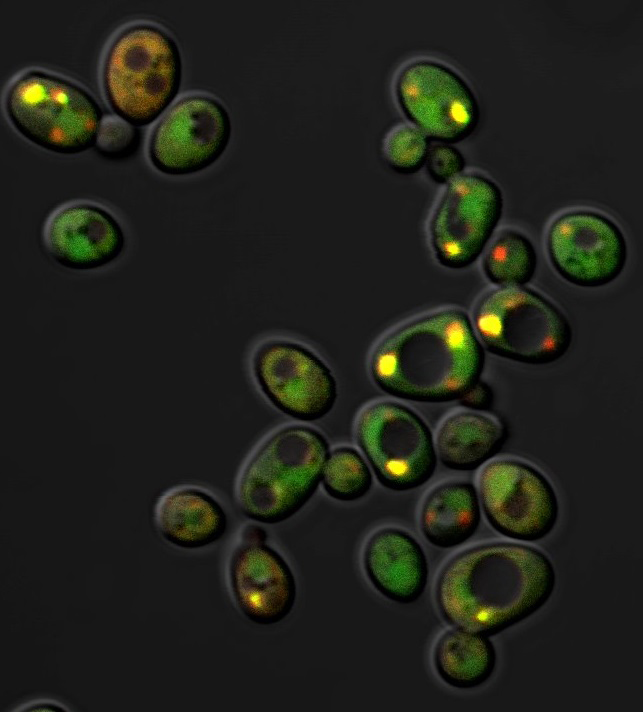
III. Genetic incompatibility and speciation
Why are there so many species in the world, and what makes them different?
Speciation generates discrete populations, which in turn play an essential role in maintaining novel adaptations during evolution. Hybrids between different species are usually inviable or sterile. One of the primary mechanisms causing postzygotic reproductive isolation is incompatibility between genes (speciation genes) from different species. These speciation genes are hypothesized to be some interacting components that cannot function properly when mixed with alleles from different species. The evolution of speciation genes is generally thought to be driven by adaptive evolution. Identifying these genes will provide more information about how speciation occurs.
Our lab uses a few closely related yeast species (the Saccharomyces sensu stricto complex) to identify genetic incompatibility leading to hybrid breakdown. By dissecting the molecular mechanisms of genetic incompatibility, we aim to know the common principles underlying yeast speciation and the driving forces behind the evolution of speciation genes.
Copyright © 2017 IMBCC. All rights reserved. |