Molecular mechanism of pre-mRNA splicing
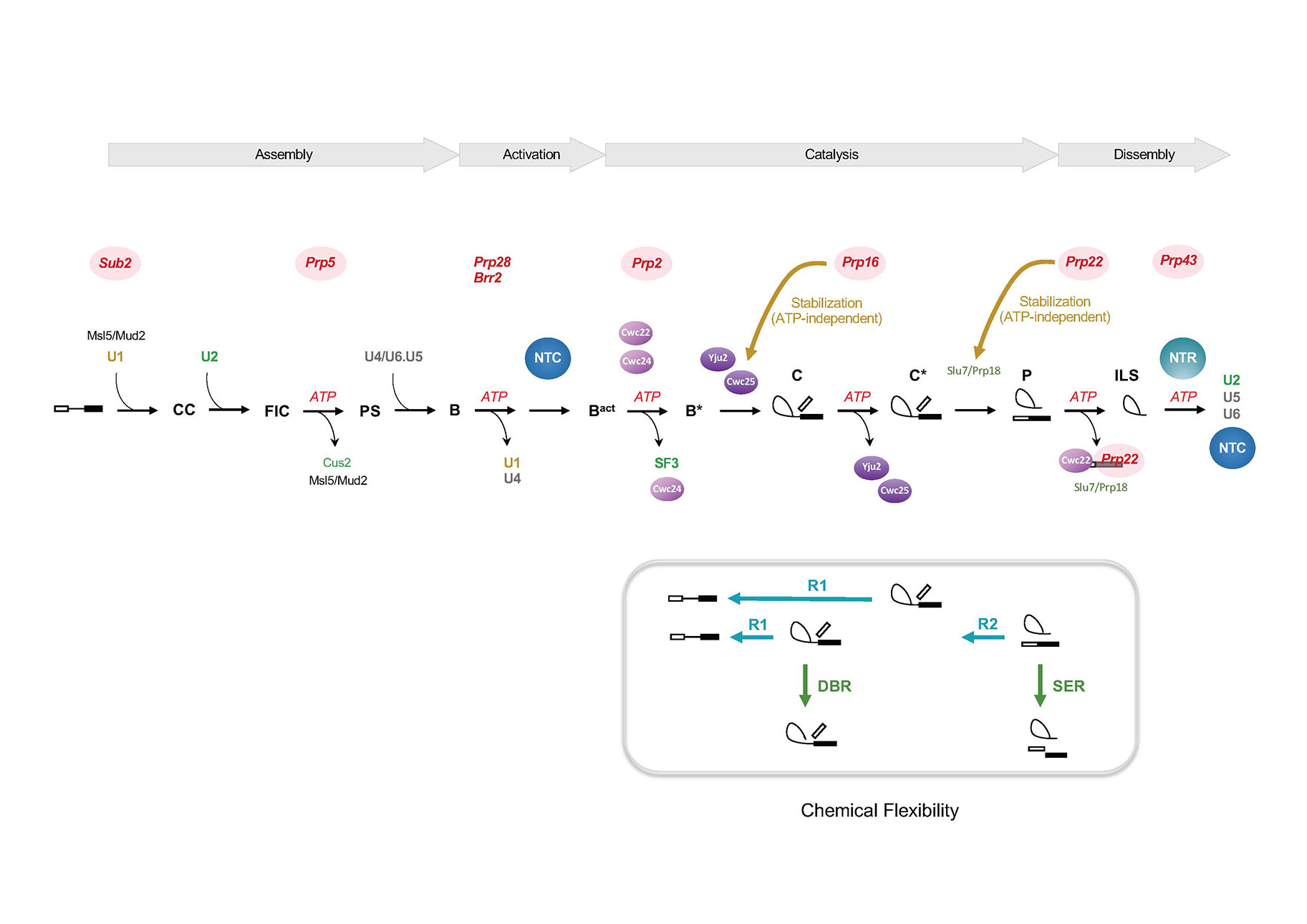
We study the molecular mechanism of the pre-mRNA splicing reaction using the budding yeast Saccharomyces cerevisiae as a model system. RNA splicing is a complex process. The reaction occurs on the spliceosome, which is assembled via ordered interactions of five snRNAs and numerous protein factors to the pre-mRNA. The snRNAs play roles in the recognition and alignment of splice sites through base-paired interactions with the intron sequence and with each other to form the catalytic core. The protein factors function to facilitate and stabilize RNA base pairings, and mediate interactions between splice sites.
Our research focuses on the functional roles of the protein splicing factors through identification and characterization of these factors using biochemical methods. We have characterized several proteins and protein complexes involved in various steps of the splicing pathway, including two protein complexes, the NTC required for spliceosome activation, and the NTR that mediates spliceosome disassembly. We have also identified proteins involved in the first catalytic step of the splicing reaction, including Yju2, Cwc22, Cwc24 and Cwc25. The identification of these factors provides further mechanistic insights underlying the catalytic reactions. We have found that the spliceosome is highly dynamic in the catalytic phase, and its conformation can readily switch between different states upon change of environmental conditions to direct different chemical reactions, much like self-splicing group II introns. While the pathway is normally guided by DExD/H-box proteins to facilitate the reaction, the purified spliceosome can catalyze different reactions autonomously without the assistance of DExD/H-box proteins under proper ionic conditions.
Eight DExD/H-box proteins participate in the splicing pathway, where they remodel the spliceosome through ATP hydrolysis. With the exception of Brr2, these proteins associate with the spliceosome only transiently and are typically not detected to associate with the spliceosome during active splicing. Several have been implicated in proofreading splice site sequences. We have further uncovered additional functions of Prp16 and Prp22 in stabilizing the binding of specific spliceosomal components in promoting catalytic reactions of mutant pre-mRNAs, thereby counteracting their canonical proofreading roles at splice sites.
- PDF, 1984-1988, Division of Biology, California Institute of Technology, USA
- PDF, 1983-1984, Laboratory of Molecular Virology, National Institutes of Health, USA
- Ph.D., 1983, Department of Biochemistry, Duke University, USA
- BS, 1977, Department of Chemistry, National Taiwan University
- Outstanding Award, National Science Council, 1994, 2010
- Hsu Yu-Ziang Award, Papers of Science and Technology, 2003
- Ho Chin-Tuei Outstanding Honorary Award in Basic Science, 2005
- Academic Award of Ministry of Education, 2007
- TWAS Prize in Biology, 2010
- Academician, Academia Sinica, Taiwan, ROC, 2012
- TWAS Member, 2013
- Outstanding Alumna of National Taiwan University, 2015
- Tarn, W.-Y., Hsu, C.-H., Huang, K.-T., Chen, H.-R., Kao, H.-Y., Lee, K.-R. and Cheng, S.-C. (1994) Functional association of essential splicing factor(s) with PRP19 in a protein complex. EMBO J. 13, 2421-2431.
- Chan, S.-P., Kao, D.-I., Tsai, W.-Y., Cheng, S.-C. (2003) The Prp19p-associated complex in spliceosome activation. Science 302: 279-282.
- Tsai, R.-T., Fu, R.-H., Yeh, F.-L., Tseng, C.-K., Lin, Y.-C., Huang, Y.-h., Cheng, S.-C. (2005) Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev. 19: 2991-3003.
- Tseng, C.-K., Cheng, S.-C. (2008) Both catalytic steps of nuclear pre-mRNA splicing are reversible. Science 320: 1782-1784. [highlighted in RNA 14, 1975-1978, 2008.]
- Tseng, C.-K., Liu, H.-L. and Cheng, S.-C. (2011) DEAH-box ATPase Prp16 has dual roles in remodeling of the spliceosome in catalytic steps. RNA 17, 145-154. [highlighted in RNA 17, 551-554, 2011.]
- Liang, W.-W., Cheng, S.-C. (2015) A novel mechanism for Prp5 in prespliceosome formation and proofreading the branch site sequence. Genes Dev. 29: 81-93.
- Chung, C.-S., Tseng, C.-K., Lai, Y.-H., Wang, H.- F., Newman, A. J., Cheng, S.-C. (2019) Dynamic interactions of proteins with pre-mRNA in mediating splicing catalysis. Nucleic Acids Res. 47: 899-910.
- Kao, C.-Y., Cao, E.-C., Wai, H. L., Cheng, S.-C. (2021) Evidence for complex dynamics during U2 snRNP selection of the intron branchpoint. Nucleic Acids Res. 49: 9965-9977.
- Chung, C.-S., Wai, H. L., Kao, C.-Y. and Cheng, S.-C. (2023) An ATP-independent role for Prp16 in promoting aberrant splicing. Nucleic Acids Res. 51, 10815–10828. [featured as NAR Breakthrough Article].
- Chung, C.-S., Tseng, C.-K., Chen, H.-C. and Cheng, S.-C. (2025) New mechanistic insights into Prp22-mediated exon ligation and mRNA release. Nucleic Acids Res. 53, gkaf823. [featured as NAR Breakthrough Article].
- Kao, C.-Y., Tsai, W.-Y., Su, Y.-L., Chung, C.-S. and Cheng, S.-C. (2025) New mechanistic insights into prespliceosome formation-Roles of Prp5 and Sub2. RNA 31, 1901-1911.