Meiosis
Meiosis is the core of Mendelian Genetics. Meiosis is a cell division process unique to the sexual reproductive cells of eukaryotes, which halves the number of chromosomes and produces haploid sperm, eggs, or fungal spores. Our laboratory uses baker’s yeast Saccharomyces cerevisiae, fission yeast Schizosaccharomyces pombe and industrial Trichoderma reesei as model organisms, employing methods from genetics, molecular biology, biochemistry, structural biology, genomics, and bioinformatics to investigate how homologous recombination (HR), DNA cytosine methyltransferase, DNA damage response and synaptonemal complex (SC) and in meiotic cells generate genetic variation and maintain genomic integrity.
The SC was first observed in Neurospora crassa by Barbara McClintock using light microscopy in 1945, and then demonstrated in 1956 in the primary spermatocytes of invertebrates and vertebrates by Montrose Moses and Don Fawcett using electron microscopy. The true biochemical roles of SC in meiosis remain controversial.
Our recent research findings and directions can be divided into three main categories:
1. The proteins (Rad51, Rad54, and Rad59) involved in HR during budding yeast meiosis, as well as the ZMM proteins (Zip1, Zip4, Mer3, and Msh4) that make up the SC, can inhibit the activity of mismatch repair (MMR). We first applied whole-genome research data and other relevant literature to explain the fallacies in the old theories in biology and genetics textbooks over the past 30 years regarding "MMR causing hybridization infertility and reproductive isolation in closely related species". Next, using the "reverse genetics" research approach, we have identified 12 HR mutant genes that specifically affect the meiosis of intraspecies SK1/S288c hybrid zygotes. The same mutant genes do not affect the meiosis of SK1/Sk1 purebred zygotes, demonstrating that multiple HR proteins can inhibit MMR activity and tolerate mismatched DNA bases during meiotic recombination. This new discovery is important for two reasons: first, it challenges the old theory that “HR is the mechanism for repairing damaged DNA with the highest fidelity”. Second, The SC has been known for nearly 70 years, but its true physiological function remains unknown. We demonstrated that the SC is responsible for suppressing MMR during meiosis, promoting recombination and exchange between homologous chromosomes with different DNA sequences, and increasing the genetic diversity of gametes or fungal sexual spores.
2. In most sexually reproducing eukaryotes (such as yeast, basidiomycetes, fruit flies, nematodes, higher plants, and mammals), meiotic cells utilize the Spo11 protein to generate DNA double-strand breaks (DSBs), thereby initiating a genetic recombination reaction. We discovered that Trichoderma reesei with the spo11 gene removed can still undergo normal meiosis and produce sexual spores. Our aim is to reveal the molecular mechanisms by which Trichoderma reesei initiates Spo11-independent meiotic recombination.
We discovered that Trichoderma reesei DNA methyltransferases have multiple functions in meiosis. First, they are responsible for initiating meiosis. Second, they promote DNA repair during meiotic recombination, thereby promoting new genetic variations. Third, through a special epigenetic mechanism, they methylate a large amount of cytosine on the parental DNA, inducing point mutations to thymine, rapidly increasing the genomic diversity of the meiotic products (sexual spores). Excessive genetic variation has a double-edged effect. While it can sometimes enhance the environmental adaptability of offspring, it often leads to genomic instability, causing sexual spores to fail to germinate into mycelia, or the newly germinated mycelia to fail to undergo normal vegetative growth. Our results can explain why most Trichoderma wild-isolates are female- or male-infertile. They can only reproduce asexually through vegetative growth of mycelium, which actually helps maintain genomic stability and perpetuate their existing survival advantages.
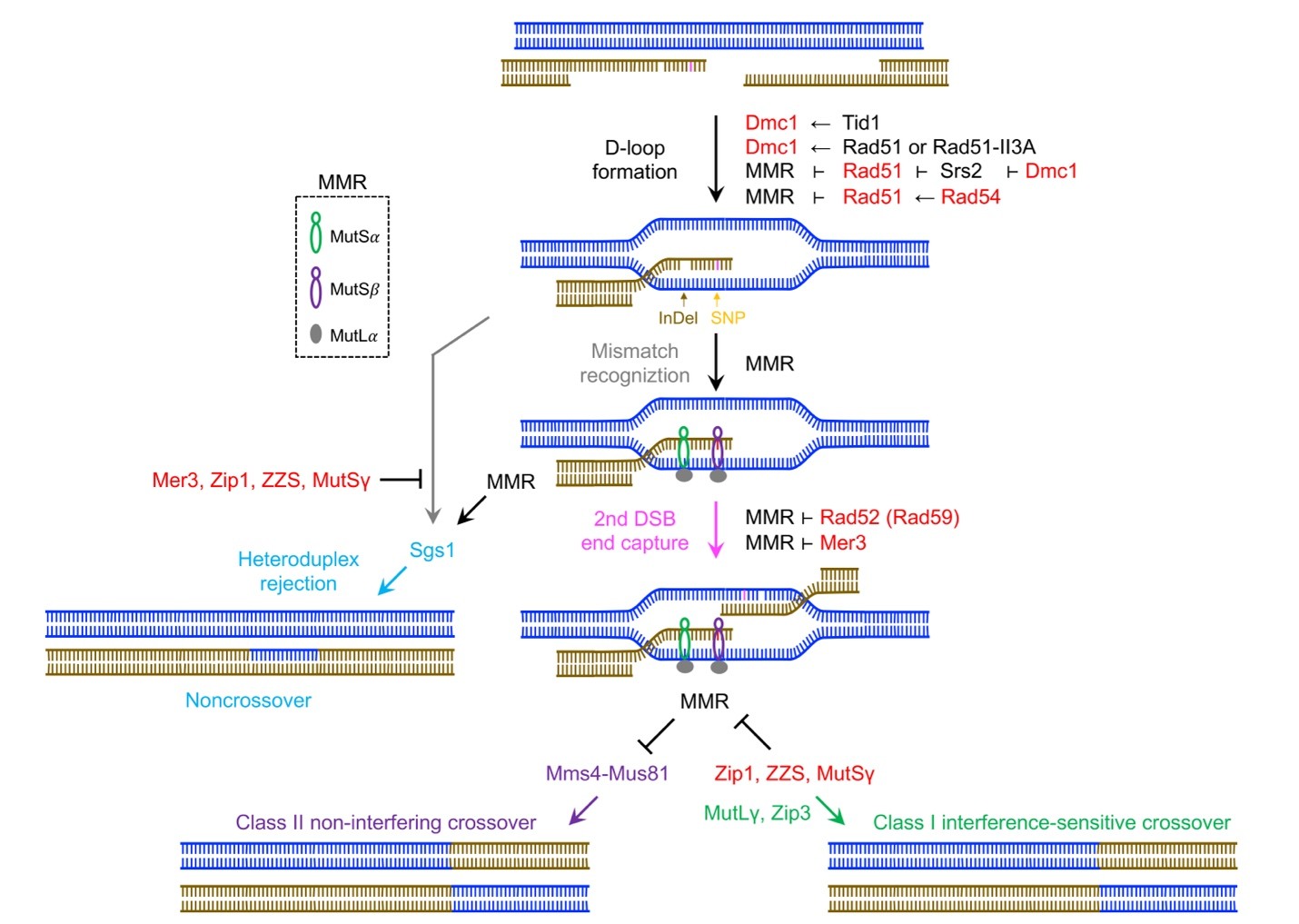
During homeologous recombination, homologous recombination (HR) proteins antagonize the mismatch repair (MMR) system in several homology-directed DSB repair steps, including D-loop formation, capture of the second processed DSB ends, and formation of ZZM-dependent class I CO products. The class II CO pathway is inhibited by MMR in hybrid zygotes. Sgs1, a primary regulator of recombination pathway choice during meiosis , can function in both MMR-dependent and -independent manners to facilitate NCO formation. Note that only one sister chromatid of each homolog and three possible interhomolog recombination products are shown. All proteins with anti-MMR activities are indicated in red. MutSα, MutSβ, and MutLγ recognize and repair mismatched base pairs in the heteroduplex DNA. Rad51 and Dmc1 utilize different mechanisms to tolerate mismatched base pairs in heteroduplex DNA and to antagonize the MMR system, respectively. Dmc1 and Tid1 (Rdh54) do not possess anti-MMR activities and Dmc1 can tolerate mismatched base pairs in heteroduplex DNA. The main function of Srs2 (a DNA helicase that promotes genome stability by dismantling toxic DNA recombination intermediates.) in HR is to disassemble the Rad51-ssDNA nucleoprotein filaments. Dmc1 is a potent inhibitor of Srs2, so it is also an anti-MMR protein.
- PDF, 1999-2000, Dept. Mol. & Cell Biol., Harvard Univ. USA
- Ph.D., 1998, Dept. Mol. & Cell Biol., Harvard Univ. USA
- MS, 1990, Inst. Life Science., Natl. Tsing Hua Univ.
- BS, 1988, Dept. Chemistry, Natl. Taiwan Univ.
- Ting-Fang Wang# and Ji-Long Liao. (2025) Homologous recombination counteracts mismatch repair to promote fertility and genetic diversity. Trends in Genetics (PMID: 41241584; DOI: 10.1016/j.tig.2025.10.009)
- Ya-Ling Hung, Chi-Ning Chuang, Hong-Xiang Kim, Hou-Cheng Liu, Jhong-Syuan Yao, Lavernchy Jovanska, Yi-Ping Hsueh, Ruey-Shyang Chen, and Ting-Fang Wang# (2025) Rad51, Rad54, and ZMM proteins antagonize the mismatch repair system to promote fertility of budding yeast intraspecies hybrid zygotes. Nucleic Acids Research (PMID: 40902007; DOI: 10.1093/nar/gkaf847)
- Hsiao-Tang Hu, Ueh-Ting Tim Wang, Bi-Chang Chen, Yi-Ping Hsueh#, Ting-Fang Wang# (2025) Ki-67 and CDK1 control the dynamic association of nuclear lipids with mitotic chromosomes. Journal of Lipid Research ( PMID: 39706365; DOI: 10.1016/j.jlr.2024.100731)
- Lavernchy Jovanska, I-Chen Lin, Jhong-Syuan Yao, Chia-Ling Chen, Hou-Cheng Liu, Wan-Chen Li, Yu-Chien Chuang, Chi-Ning Chuang, Albert Chen-Hsin Yu, Hsin-Nan Lin, Wen-Li Pong, Chang-I Yu, Ching-Yuan Su, Yi-Ping Chen, Ruey-Shyang Chen, Yi-Ping Hsueh, Hanna S Yuan, Ljudmilla Timofejeva, Ting-Fang Wang# (2024) DNA cytosine methyltransferases differentially regulate genome-wide hypermutation and interhomolog recombination in Trichoderma reesei meiosis. Nucleic Acids Research (PMID: 39021337; DOI: 10.1093/nar/gkae611)
- Chi-Ning Chuang, Hou-Cheng Liu, Tai-Ting Woo, Ju-Lan Chao, Chiung-Ya Chen, Hisao-Tang Hu, Yi-Ping Hsueh, Ting-Fang Wang# (2024) Noncanonical usage of stop codons in ciliates expands proteins with structurally flexible Q-rich motifs. eLife (PMID: 38393970; DOI: 10.7554/eLife.91405)
- Chia-Lin Chen, Wan-Chen Li, Yu-Chien Chuang, Hou-Cheng Liu, Chien-Hao Huang, Ko-Yun Lo, Chung-Yu Chen, Fang-Mo Chang, Guo-An Chang, Yu-Ling Lin, Wen-denr Yang, Ching-Hua Su, Tsung-Ming Yeh, and Ting-Fang Wang# (2022) Sexual crossing, chromosome-level genome sequences, and comparative genomic analyses for the medicinal mushroom Taiwanofungus camphoratus (Syn. Antrodia cinnamomea; Antrodia camphorata). Microbiology spectrum (PMID: 35196809; DOI: 10.1128/spectrum.02032-21)
- Wan-Chen Li, Ting-Chan Lin, Chia-Ling Chen, Hou-Cheng Liu, Hisn-Nan Lin, Ju-Lan Chao, Cheng-Hsilin Hsieh, Hui-Fang Ni, Ruey-Shyang Chen# and Ting-Fang Wang# (2021) Complete genome sequences and genome-wide characterization of Trichoderma biocontrol agents provide new insights into their evolution and variation in genome organization, sexual development, and fungal-plant interactions. Microbiology spectrum (PMID: 34908505; DOI: 10.1128/Spectrum.00663-21)
- Wan-Chen Li, Chia-Yi Lee, Wei-Hsuan Lan, Tai-Ting Woo, Hou-Cheng Liu, Hsin-Yi Yeh, Hao-Yan Chang, Yu-Chien Chuang, Chi-Ning Chuang, Chia-Lin Chen, Yi-Ping Hsueh, Hung-Wen Li#, Peter Chi#, and Ting-Fang Wang# (2021) Trichoderma reesei Rad51 tolerates mismatches in hybrid meiosis with diverse genome sequences. Proceedings of the National Academy of Sciences (PMID: 33593897; DOI: 10.1073/pnas.2007192118)
- Wan-Chen Li and Ting-Fang Wang# (2021) PacBio long-read sequencing, assembly, and Funannotate reannotation of the complete genome of Trichoderma reesei QM6a. Methods in Molecular Biology (PMID: 33165795; DOI: 10.1007/978-1-0716-1048-0_21)
- Hou-Cheng Liu, Wan-Chen Li, and Ting-Fang Wang# (2021) TSETA: A third-generation sequencing-based computational tool for mapping and visualization of SNPs, meiotic recombination products, and RIP Mutations. Methods in Molecular Biology (PMID: 33165796; DOI: 10.1007/978-1-0716-1048-0_2)
- Tai-Ting Woo, Chi-Ning Chuang, Mika Higashide, Akira Shinohara, and Ting-Fang Wang# (2020) Dual roles of yeast Rad51 N-terminal domain in repairing DNA double-strand breaks. Nucleic Acids Research (PMID: 32652040; DOI: 10.1093/nar/gkaa587)
- Wan-Chen Li, Hou-Cheng Liu, Ying-Jyun Lin, Shu-Yun Tung, Ting-Fang Wang# (2020) Third-generation sequencing-based mapping and visualization of single nucleotide polymorphism, meiotic recombination, illegitimate mutation and repeat-induced point mutation. NAR Genomics and Bioinformatics (PMID: 33575607; DOI: 10.1093/nargab/lqaa056)
- Wang, T.-F., Chin-Ning Chuang and Tai-Ting Woo. “Method and vector for enhancing protein expression”. US provisional application (63/036,704), Patent Cooperation Treaty application (PCT/US2021/036247) and Taiwan application (110120541).
- Wang, T.-F., “Method for producing segmental aneuploidy (SAN) strains of Trichoderma reesei via sexual crossing and SAN strains produced therefrom”. Patent Cooperation Treaty (PCT/US14/58654) and US (9598738B2), Taiwan (I570236) and People’s Republic of China (IA001/ACA081CN).
- Wang, T. F. and Yang, S.M. “Methods of producing virus-like particles of piconavirus using a small-ubiquitin-related fusion protein expression system”. US (8663951B2) and Taiwan (I415623).
- Wang, T.-F. “SUMO fusion protein expression system for producing native proteins”. USA 8034910BZ.