剪接反應分子機制的探討
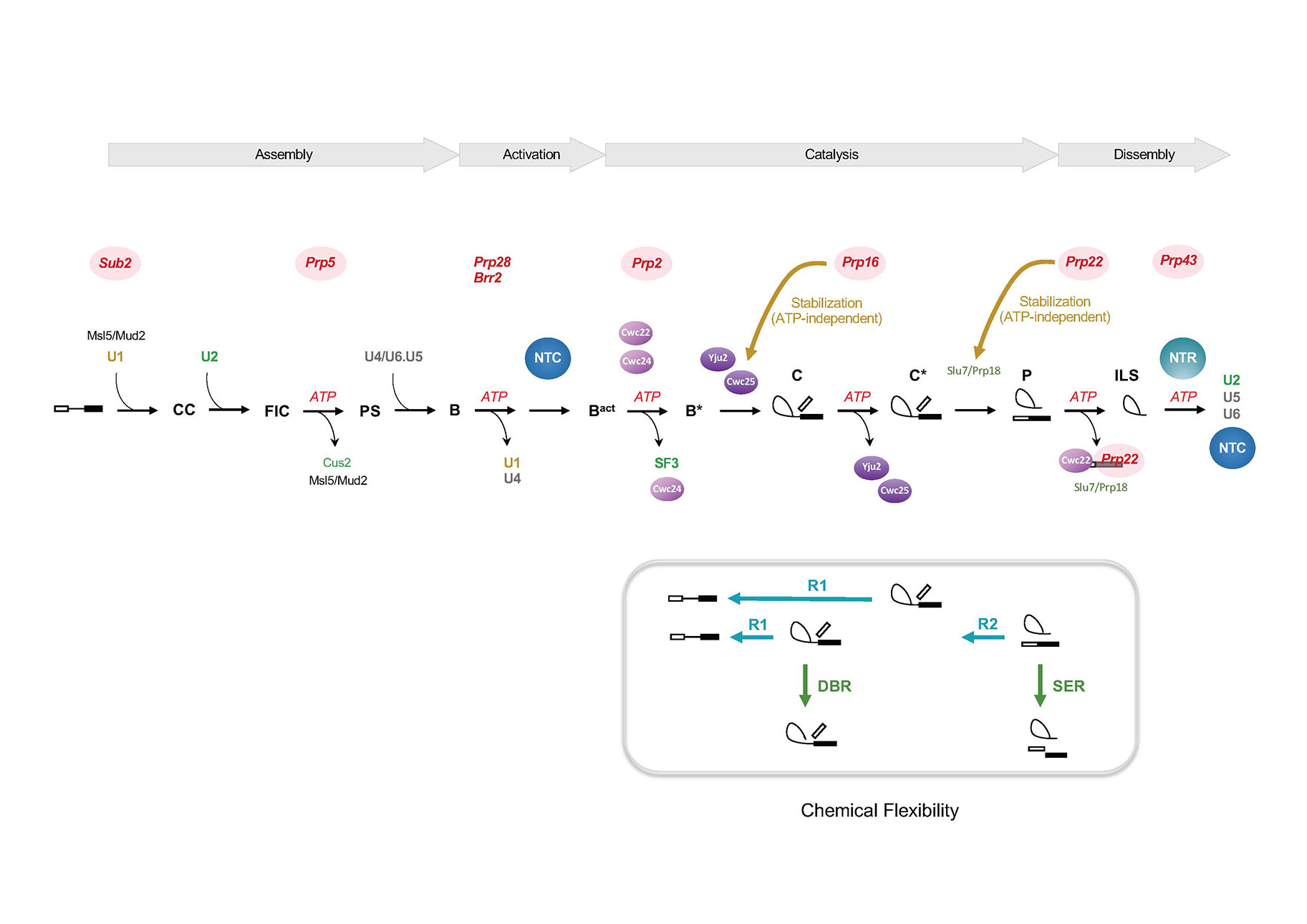
我們的研究是以酵母菌為模式系統探討訊息核醣核酸剪接反應的分子機制。剪接反應在剪接體上進行,反應步驟非常複雜。剪接體是由五個小核核醣核酸及許多蛋白因子依序結合到前訊息核醣核酸上組合而成。小核核醣核酸藉由鹼基配對辨識剪接位並促進剪接位對位以形成剪接反應中心,蛋白因子則穩定這些鹼基配對,並協助剪接位的連結。
我們的研究著重於探討剪接蛋白因子在剪接反應中的功能。實驗方法主要利用生化分析,鑑定新的剪接因子,研究其分子特性,了解其功能。我們過去鑑定了數個剪接蛋白因子及蛋白複合體,並對其功能作詳細分析。這其中包括兩個蛋白複合體,分別參與在剪接體活化及剪接體拆解,以及多個現參與在第一步的剪接催化反應的蛋白因子,包括Yju2、Cwc22、Cwc24 和 Cwc25。發掘新的因子,讓我們對剪接體組合的路徑能作更詳盡的分析,並可分離出剪接體的中間產物以為進一步的研究。我們發現剪接體呈高度的動態狀態,分離出的剪接體,其周圍離子環境會影響其構型改變以主導其催化的化學反應,此現象與第二型自發性剪接反應的介入子相類似。
正常的剪接反應需要八個DExD/H-box蛋白,它們利用水解ATP來促進剪接體組成轉換、構形轉變以加速反應的進行。除了Brr2外,它們在正常反應時只與剪接體有短暫的交互作用,在剪接體上不會偵測到它們的存在。其中有幾個成員曾被舉證認為可控制剪接反應準確度。我們發現Prp16和Prp22另具有不依賴ATP的功能,可以穩定剪接因子與剪接體的結合以促進剪接反應的進行,對於剪接位有突變的pre-mRNA形成錯誤的反應生成物。這個發現與這些蛋白具有控制剪接反應準確度的角色相違背。
- PDF, 1984-1988, Division of Biology, California Institute of Technology, USA
- PDF, 1983-1984, Laboratory of Molecular Virology, National Institutes of Health, USA
- Ph.D., 1983, Department of Biochemistry, Duke University, USA
- BS, 1977, Department of Chemistry, National Taiwan University
- Outstanding Award, National Science Council, 1994, 2010
- Hsu Yu-Ziang Award, Papers of Science and Technology, 2003
- Ho Chin-Tuei Outstanding Honorary Award in Basic Science, 2005
- Academic Award of Ministry of Education, 2007
- TWAS Prize in Biology, 2010
- Academician, Academia Sinica, Taiwan, ROC, 2012
- TWAS Member, 2013
- Outstanding Alumna of National Taiwan University, 2015
- Tarn, W.-Y., Hsu, C.-H., Huang, K.-T., Chen, H.-R., Kao, H.-Y., Lee, K.-R. and Cheng, S.-C. (1994) Functional association of essential splicing factor(s) with PRP19 in a protein complex. EMBO J. 13, 2421-2431.
- Chan, S.-P., Kao, D.-I., Tsai, W.-Y., Cheng, S.-C. (2003) The Prp19p-associated complex in spliceosome activation. Science 302: 279-282.
- Tsai, R.-T., Fu, R.-H., Yeh, F.-L., Tseng, C.-K., Lin, Y.-C., Huang, Y.-h., Cheng, S.-C. (2005) Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev. 19: 2991-3003.
- Tseng, C.-K., Cheng, S.-C. (2008) Both catalytic steps of nuclear pre-mRNA splicing are reversible. Science 320: 1782-1784. [highlighted in RNA 14, 1975-1978, 2008.]
- Tseng, C.-K., Liu, H.-L. and Cheng, S.-C. (2011) DEAH-box ATPase Prp16 has dual roles in remodeling of the spliceosome in catalytic steps. RNA 17, 145-154. [highlighted in RNA 17, 551-554, 2011.]
- Liang, W.-W., Cheng, S.-C. (2015) A novel mechanism for Prp5 in prespliceosome formation and proofreading the branch site sequence. Genes Dev. 29: 81-93.
- Chung, C.-S., Tseng, C.-K., Lai, Y.-H., Wang, H.- F., Newman, A. J., Cheng, S.-C. (2019) Dynamic interactions of proteins with pre-mRNA in mediating splicing catalysis. Nucleic Acids Res. 47: 899-910.
- Kao, C.-Y., Cao, E.-C., Wai, H. L., Cheng, S.-C. (2021) Evidence for complex dynamics during U2 snRNP selection of the intron branchpoint. Nucleic Acids Res. 49: 9965-9977.
- Chung, C.-S., Wai, H. L., Kao, C.-Y. and Cheng, S.-C. (2023) An ATP-independent role for Prp16 in promoting aberrant splicing. Nucleic Acids Res. 51, 10815–10828. [featured as NAR Breakthrough Article].
- Chung, C.-S., Tseng, C.-K., Chen, H.-C. and Cheng, S.-C. (2025) New mechanistic insights into Prp22-mediated exon ligation and mRNA release. Nucleic Acids Res. 53, gkaf823. [featured as NAR Breakthrough Article].
- Kao, C.-Y., Tsai, W.-Y., Su, Y.-L., Chung, C.-S. and Cheng, S.-C. (2025) New mechanistic insights into prespliceosome formation-Roles of Prp5 and Sub2. RNA 31, 1901-1911.