Structural Insights into RNA and DNA Metabolism
We investigate RNA- and DNAbinding proteins that play key roles in nucleic acids metabolism. The overall goal is to discover the structure-based mechanism of these proteins in nucleic acids recognition, processing, modification and degradation. Using the main tool of X-ray crystallography, together with biochemical and biophysical approaches, we have shown how a group of proteins function in trimming, unwinding, degradation and modification of RNA/DNA in diverse ways.
Over expression, mutations and misfolding of most of the proteins that we study are linked to human diseases, ranging from neurodegenerative disorders to inflammatory diseases. Therefore, understanding how these proteins recognize RNA/DNA and participate in nucleic acids metabolism and how they lose their functions not only advance our knowledge in the working mechanisms of RNA/DNA metabolism, but also pave the way for strategy development of disease treatments.
- PDF, 1988-1992, Molecular Biology Institute UCLA, USA.
- Ph.D., 1988, Department of Chemistry University of Southern California, USA.
- BS, 1983, Department of Chemistry Tunghai University
- 1998, Academia Sinica Early-Career Investigator Research Achievement Award
- 1998, 2003, 2011, Research Excellence Award, National Science and Technology Council
- 2006-2010, 2011-2015, 2016-2000, 2021-2025, Academia Sinica Investigator Award
- 2020, Academic Award of Ministry of Education
- Hsiao, Y.-Y., Fang, W.-H., Lee, C.-C, Chen, Y.-P., Hsu, P.-C., Yuan, H. S. (2014) Structural insights into DNA repair by RNase T, an exonuclease processing 3′ end of structured DNA in repair pathways. PLoS Biol. 12: e1001803.
- Lin, J. L. J., Nakagawa, A., Skeen-Gaar, R., Yang, W.-Z., X., Ge, S., Mitani, Xue*, D, Yuan, H. S. (2016) Oxidative stress impairs cell death by repressing the nuclease activity of Endonuclease G. Cell Rep., 16: 279-287.
- Lin, J.L.J., Wu, C.C., Yang, W.-Z., Yuan, H. S. (2016) Crystal structure of endonuclease G in complex with DNA reveals how it nonspecifically degrades DNA as a homodimer. Nucleic Acids Res. 44: 10480- 10490.
- Huen, J., Lin, C.-L., Yi, W.-L., W.-Z. Yang, Golzarroshan, B., Yuan, H. S. (2017) Structural insights into a unique dimeric DEAD-box helicase CshA that promotes RNA decay. Structure 25: 469–481.
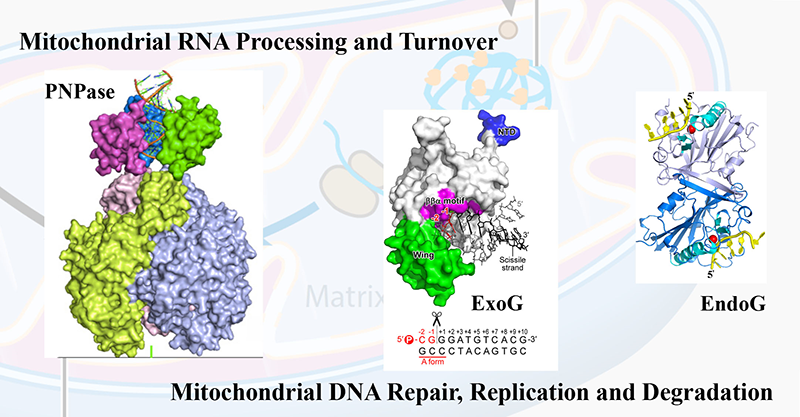
- Golzarroshan, B., Lin, C.-L., Li, C.-L., Yang, W.-Z., Chu, L.-Y., Agrawal, S., Yuan, H. S. (2018) Crystal structure of dimeric human PNPase reveals why disease-linked mutants suffer from low RNA import and degradation activities. Nucleic Acids Res. 46: 8630-8640.
- Wu, C.C., Lin, J.L.J., Yang-Yen, H.-F., Yuan, H. S. (2019) A unique exonuclease ExoG cleaves at the junction between RNA and DNA in mitochondrial DNA replication. Nucleic Acids Res. 47: 5405-5419.
- Lin, C.C., Chen, Y. P., Yang, W. Z., Shen, J. C. K., Yuan, H. S. (2020) Structural insights into CpG-specific DNA methylation by human DNA methyltransferase 3B. Nucleic Acids Res. 48: 3949-3961.
- Wu, C.C., Lin, J.L.J., Yuan, H. S. (2020) Structures, mechanisms and functions of His-Me finger nucleases. Trends Biochem. Sci. 45: 935-946.