General Principles and Molecular Mechanisms of Endosymbiosis, Phenotypic Robustness, and Speciation
Genomic Analysis of Endosymbiosis
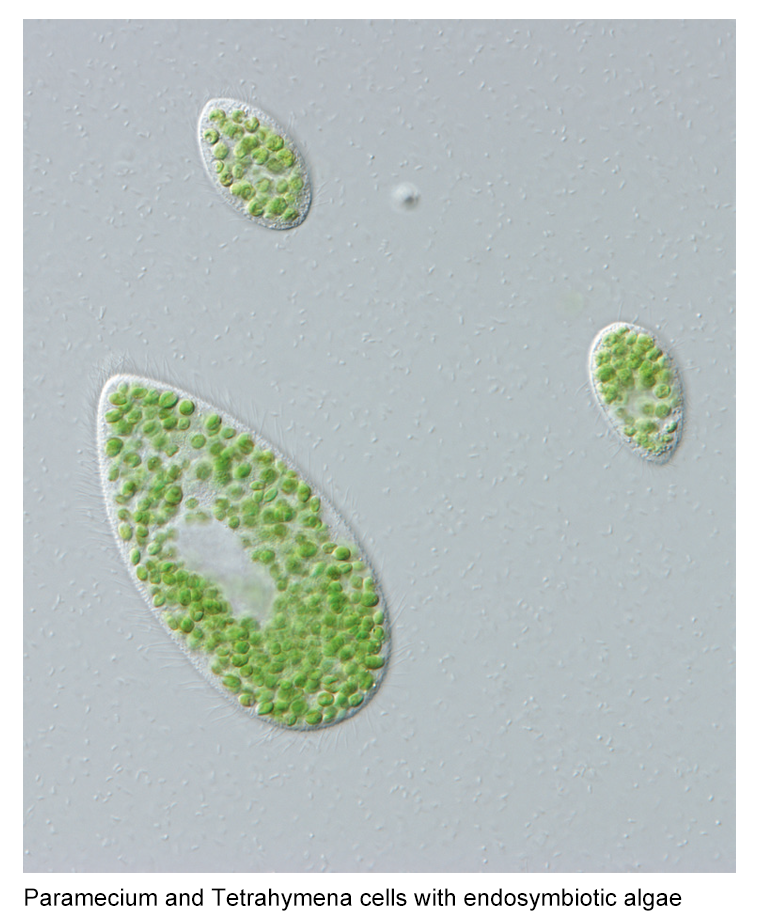
Why can an organism live inside another organism without causing trouble? Endosymbiosis is one of the major forces driving the evolution of eukaryotic cells, which has occurred multiple times in different lineages during evolutionary history. However, the initial process of how the endosymbiosis is formed and established in the host cell is still unclear. We use the ciliates and the algae to study the early stage of endosymbiosis. Ciliates are an ancient and diverse group of protists exhibiting extraordinary features of cell biology. The green algae have been utilized in many industrial applications such as biofuel production. These two systems allow us to explore the genetic basis of endosymbiosis establishment and also other aspects of biology.
Molecular Mechanisms and the Evolution of Phenotypic Robustness
Why can the cell maintain consistent performance under a variety of conditions and genetic backgrounds? The ability to maintain a stable phenotype despite environmental and genetic perturbations is a fundamental property of living systems. It has been speculated that cells contain various buffering systems that can establish phenotypic robustness under general conditions and regulate it under stress. Buffering systems are crucial in stabilizing cell physiology and developmental processes from environmental and genetic perturbations. Besides, buffering systems may play an important role in setting the tempo of evolution since they can moderate these perturbations or help a population accumulate genetic variations essential for selection. Although the concept of genetic and non-genetic buffering is well established, the underlying mechanisms are largely unclear. In our lab, a systematic approach combining experimental evolution, genomics, and proteomics analyses is used to understand their molecular basis.
- PDF, 2000-2006, Dept. Dept. Mol. & Cell Biol., Harvard Univ., USA
- Ph.D., 1999, Dept. Mol. Cell, & Dev. Biol., Yale Univ., USA
- MS, 1990, Dept. Life Sciences, Natl. Tsing-Hua Univ.
- BS, 1988, Dept. Chemical Engineering, Natl. Tsing-Hua Univ.
- 2022, Ministry of Education Annual Academic Award
- 2021, Ministry of Science and Technology Outstanding Research Award
- 2018, Outstanding Scholar Award, Foundation for the Advancement of Outstanding Scholarship
- 2013, National Science Council Outstanding Research Award
- 2009, Academia Sinica Research Award for Junior Investigators in Taiwan
- 2007, International Human Frontier Science Program (HFSP) Young Investigators’ Award
- Kamal, M.M., Cheng, Y.-H., Chu, L.-W., Nguyen, P.T., Liu, C.-F.J., Liao, C.-W., Posch, T., and Leu, J.-Y.* “Environment-dependent mutualism-parasitism transitions in the incipient symbiosis between Tetrahymena utriculariae and Micractinium tetrahymenae” ISME J. 19(1): wraf203 (2025).
- Hsu, P.-C.*, Lu, T.-C., Hung, P.-H. and Leu, J.-Y.* “Protein moonlighting by a target gene dominates phenotypic divergence of the Sef1 transcription regulatory network in yeasts.” Nucleic Acids Res. 52(22): 13914-13930 (2024).
- Yeh, C.-W., Hsu, K.-L., Lin, S.-T., Huang, W.-C., Yeh, K.-H., Liu, C.-F.J., Wang, L.-C., Li, T.-T., Chen, S.-C., Yu, C.-H., Leu, J.-Y., Yeang, C.-H., Yen, H.-C.S.* “Altered assembly paths mitigate interference among paralogous complexes.” Nature Commun. 15:7169 (2024).
- Lin, W.-H., Opoc, F.J.G., Liao, C.-W., Roy, K.R., Steinmetz, L.M. and Leu, J.-Y.* “Histone deacetylase Hos2 regulates protein expression noise by potentially modulating the protein translation machinery.” Nucleic Acids Res. 52(13): 7556-7571 (2024).
- Chen, Y.-H., Chao, K.-H., Wong, J.-Y., Liu, C.-F.J., Leu, J.-Y.* and Tsai, H.-K.*, “A feature extraction free approach for protein interactome inference from co-elution data.” Brief. Bioinform. 24(4): bbad229 (2023).
- Lai, H.-Y., Yu, Y.-H., Jhou, Y.-T., Liao, C.-W. and Leu, J.-Y.* “Multiple intermolecular interactions facilitate rapid evolution of essential genes.” Nature Ecol. Evol. 7(5): 745-755 (2023).
- Swamy, K.B.S., Lee, H.-Y., Ladra, C., Liu, C.-F.J., Chao, J.-C., Chen, Y.-Y. and Leu, J.-Y.* “Proteotoxicity caused by perturbed protein complexes underlies hybrid incompatibility in yeast.” Nature Communications 13: 4394 (2022).
- Amine, A.A.A., Liao, C.-W., Hsu, P.-C., Opoc, F.J.G. and Leu, J.-Y.*, “Experimental evolution improves mitochondrial genome quality control in Saccharomyces cerevisiae and extends its replicative lifespan.” Current Biology 31(16): 3663-3670 (2021).
- Leu, J.-Y., Chang, S.-L., Chao, J.-C., Woods, L.C. and McDonald, M.J.* “Sex alters molecular evolution in diploid experimental populations of S. cerevisiae.” Nature Ecology Evolution 4(3): 453-460 (2020).
- You, S.-T., Jhou, Y.-T., Kao, C.-F. and Leu, J.-Y.* “Experimental evolution reveals a general role for the methyltransferase Hmt1 in noise buffering.” PLoS Biology 17(10): e3000433 (2019).
- McDonald, M.J.*, Yu, Y.-H., Guo, J.-F., Chong, S.-Y., Kao, C.-F. and Leu, J.-Y.* “Mutation at a distance caused by homopolymeric guanine repeats in Saccharomyces cerevisiae.” Science Advances 2: e1501033 (2016).
- Siegal, M.L.* and Leu, J.-Y.* “On the nature and evolutionary impact of phenotypic robustness mechanisms.” Annual Review of Ecology, Evolution, and Systematics 45: 495-517 (2014).
- Hsieh, Y.-Y., Hung, P.-H., Leu, J.-Y.* “Hsp90 regulates non-genetic variation in response to environmental stress.” Molecular Cell 50: 82-92 (2013).
- Chou, J.-Y., Hung Y.-S., Lin, K.-H., Lee, H.-Y. and Leu, J.-Y.* “Multiple molecular mechanisms cause reproductive isolation between three yeast species.” PLoS Biology 8(7): e1000432 (2010).
- Lee, H.-Y., Chou, J.-Y., Cheong, L., Chang, N.-H., Yang, S.-Y. and Leu, J.-Y. (2008) “Incompatibility of nuclear and mitochondrial genomes causes hybrid sterility between two yeast species.” Cell 135: 1065-1073.