蛋白質中“質”與“量”的調控
蛋白質是組成生物體的主要成分及執行細胞生理功能的最小單位,因此正確的轉譯遺傳密碼對於生命現象的維持至關重要,但是細胞時常受到環境的刺激進而引發蛋白質畸變,例如遺傳密碼的突變而導致蛋白質的轉譯錯誤,折疊不當及運送錯誤。 於此同時,細胞演化出一套精準的蛋白質品管系統來維持細胞的恆定。此系統會分別對於“好”或“壞”的蛋白質進行分類,並選擇性地只消除“壞”蛋白質。而本實驗室致力於探討細胞是如何使用有限的泛素連接酶來捕獲廣泛的缺陷蛋白。 我們對於泛素連接酶是透過哪些分子機制得已區分“好”還是“壞”的蛋白質感到好奇,我們的研究目的便是解決以上生物學中最核心的問題。
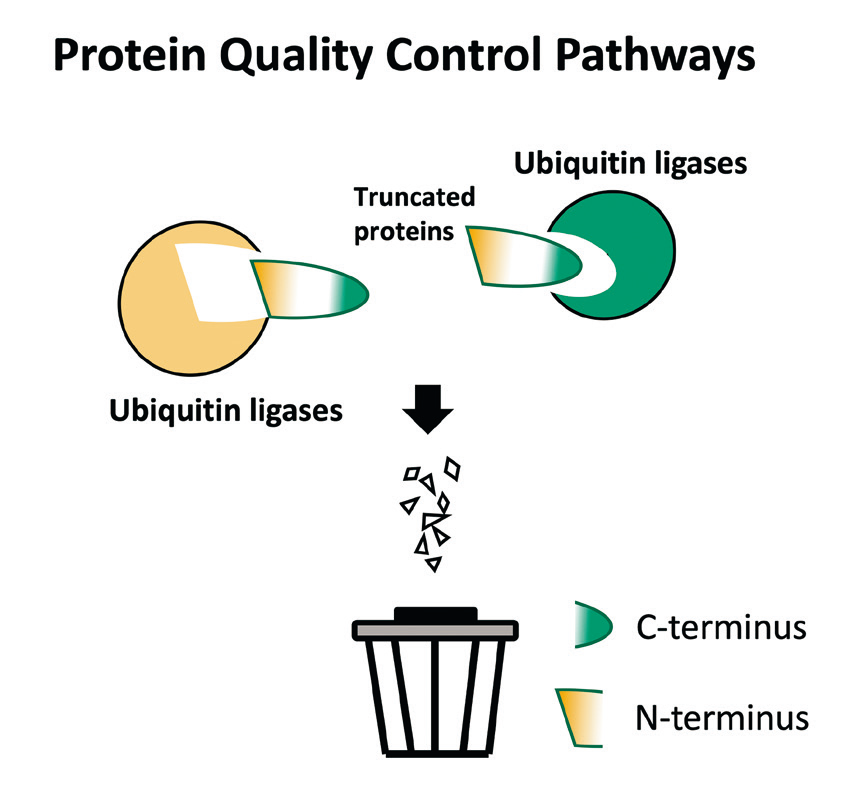
蛋白質完整性的指標其實就藏在蛋白質的末端。早在1986 年, Varshavsky 等人便發現蛋白質的壽命長短取決於N 端氨基酸序列,稱之為 N-degrons。於2015年,我們進一步揭露C端氨基酸序列 (C-degron) 對於蛋白質的壽命長短亦扮演重要角色。此研究成果無疑在蛋白降解的領域中開啟新的篇章。 C-degron 降解途徑具有以下幾個重要生物意義。1. 清除定位錯誤的蛋白質。2. 去除蛋白酶分解後的副產物與被截短的硒蛋白。3. 形塑真核動物中蛋白質體的演化。我們當前的研究重點旨在開拓 C-degron 降解途徑的其他功能。
- PDF, 2003-2010, Department of Genetics, Harvard Medical School, USA
- Ph.D., 2003, Department of Biology New York University, USA
- MS, 1996, Institute of Molecular Medicine, National Taiwan University
- BS, 1994, Dept. Department of Zoology, National Taiwan University
- 2011, 傑出人才講座
- 2011, 李氏傳統基金會獎助金
- 2012-2016, 中研院前瞻計畫
- 2019-2023, 中研院深耕計畫
- 2017-2022, 2022-2027, 科技部尖端計畫
- Yen, H.C.S., Elledge, S.J. (2008) Identification of SCF ubiquitin ligase substrates by global protein stability profiling. Science 332: 923-929.
- Yen, H.C.S., Xu, Q., Chou, D.M., Zhao, Z., Elledge, S.J. (2008) Global protein stability profiling in mammalian cells. Science 332: 918-923.
- Emanuele, M.J., Elia, A.E.H., Xu, Q., Thoma, C.R., Izhar, L., Leng, Y., Guo, A., Chen, Y.N., Rush, J., Hsu, W.C., Yen, H.C.S., Elledge, S.J. (2011) Global identification of modular Cullin-RING ligase substrates. Cell 147: 459- 474.
- Lin, H.C., Ho, S.C., Chen, Y.Y., Khoo, K.H., Khoo, K.H., Hsu, P.H., Yen, H.C.S. (2015) CRL2 aids elimination of truncated selenoproteins produced by failed UGA/Sec decoding. Science 349: 91-95.
- Chen, Y.F., Lin, H.C., Chuang, K.N., Lin, C.H., Yen, H.C.S., Yeang, C.H. (2017) A quantitative model for the rate- limiting process of UGA alternative assignments to stop and selenocysteine codons. PLoS Comput. Biol. 13: e1005367.
- Lin, H.C., Yeh, C.H., Chen, Y.F., Lee, T.T., Hsieh, P.Y., Rusnac, D.V., Lin, S.Y., Elledge, S.J., Zheng, N., Yen, H.C.S. (2018) C-terminal end-directed protein elimination by CRL2 ubiquitin ligases. Mol. Cell 70: 602- 613.
- Rusnac, D.V., Lin, S.C., Canzani, D., Tien, K., Hinds, T.R., Tsue, A.F., Bush, M.F., Yen, H.C.S., Zheng, N. (2018) Recognition of diglycine C-end degron by CRL2KLHDC2 ubiquitin ligase. Mol. Cell 72: 813-822.
- Yeh, C.W., Huang, W.C., Hsu, P.H., Yeh, K.H., Wang, L.C., Hsu, W.H., Lin, H.C., Chen, Y.N., Chen, S.C., Yeang, C.H., Yen, H.C.S. (2021) The C-degron pathway eliminates mislocalized proteins and products of deubiquitinating enzymes. EMBO J. 40: e105846.
- Hsu, Kuan-Lun, Yen, Hsueh-Chi S., and Yeang, Chen-Hsiang (2022). Cooperative stability renders protein complex formation more robust and controllable. Sci Rep 12, 10490.
- Yeh, C. W., Hsu, K. L., Lin, S. T., Huang, W. C., Yeh, K. H., Liu, C. J., Wang, L. C., Li, T. T., Chen, S. C., Yu, C. H., Leu, J. Y., Yeang, C. H., & Yen, H. S. (2024). Altered assembly paths mitigate interference among paralogous complexes. Nature communications, 15(1), 7169.