Protein Quality Control
Proteome fidelity is of critical importance in almost all cellular processes, yet it is constantly challenged by a diversity of protein aberrations arising from genetic mutations, erroneous transcription and translation, improper folding, and damage induced by various environmental stresses. A protein quality control system must sort through unlimited variations representing “bad” or “good” proteins and selectively eliminate the “bad” proteins. How can such a wide spectrum of defective proteins be captured using limited inspectors? Which molecular characteristics distinguish proteins directed to the “good” pool versus the “bad” pool? Our study addresses these long-standing fundamental questions, which have a broad impact on biology.
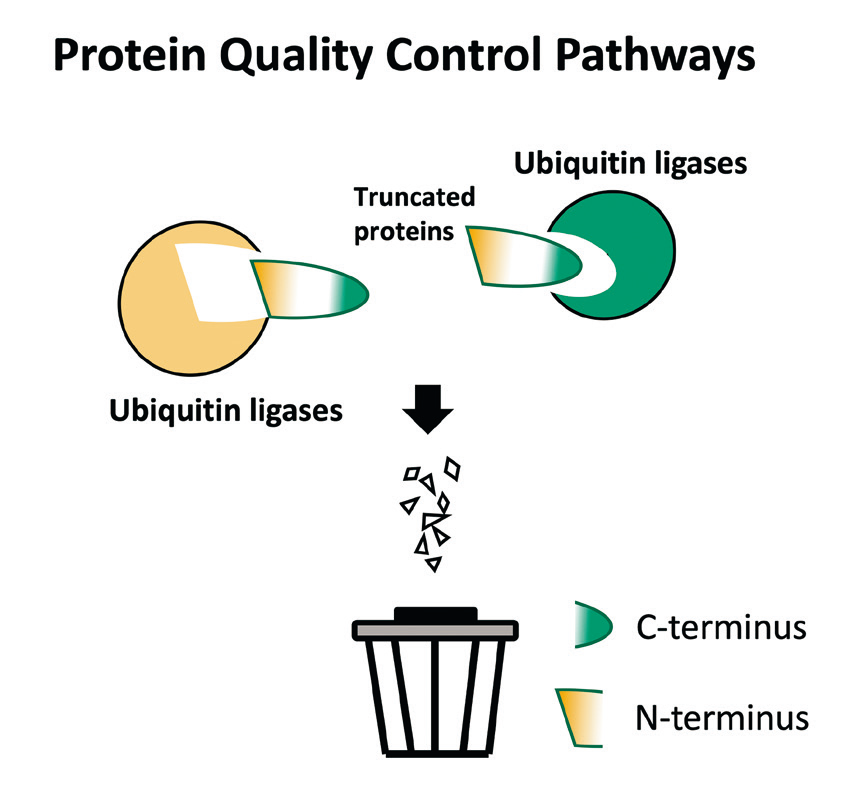
Protein termini are indicators of protein integrity. Discovered by Varshavsky et al. in 1986, N-degrons were the first degradation signals to be identified in short-lived proteins. In 2015, we opened a second chapter in “end-mediated” proteolysis by characterizing a C-degron pathway, by which proteins with illegitimate C-termini are eliminated. We uncovered the role of the C-degron pathway in clearing mislocalized proteins, products of proteases, and truncated selenoproteins due to translation errors. Importantly, we have provided evidence supporting that avoidance of C-degron-mediated degradation shapes eukaryotic protein evolution/selection. Accordingly, C-terminal-specific recognition for subsequent protein degradation is likely more prevalent than heretofore acknowledged. Our current research aims to identify additional functions of the C-degron pathway.
- PDF, 2003-2010, Department of Genetics, Harvard Medical School, USA
- Ph.D., 2003, Department of Biology New York University, USA
- MS, 1996, Institute of Molecular Medicine, National Taiwan University
- BS, 1994, Dept. Department of Zoology, National Taiwan University
- 2011, Outstanding Scholar Awards
- 2011, Li Foundation Heritage Prize
- 2012-2016, Academia Sinica Career Development Award
- 2019-2023, Academia Sinica Investigator Award
- 2017-2022, 2022-2027, Frontier Science Grant, National Science and Technology Council
- Yen, H.C.S., Elledge, S.J. (2008) Identification of SCF ubiquitin ligase substrates by global protein stability profiling. Science 332: 923-929.
- Yen, H.C.S., Xu, Q., Chou, D.M., Zhao, Z., Elledge, S.J. (2008) Global protein stability profiling in mammalian cells. Science 332: 918-923.
- Emanuele, M.J., Elia, A.E.H., Xu, Q., Thoma, C.R., Izhar, L., Leng, Y., Guo, A., Chen, Y.N., Rush, J., Hsu, W.C., Yen, H.C.S., Elledge, S.J. (2011) Global identification of modular Cullin-RING ligase substrates. Cell 147: 459- 474.
- Lin, H.C., Ho, S.C., Chen, Y.Y., Khoo, K.H., Khoo, K.H., Hsu, P.H., Yen, H.C.S. (2015) CRL2 aids elimination of truncated selenoproteins produced by failed UGA/Sec decoding. Science 349: 91-95.
- Chen, Y.F., Lin, H.C., Chuang, K.N., Lin, C.H., Yen, H.C.S., Yeang, C.H. (2017) A quantitative model for the rate- limiting process of UGA alternative assignments to stop and selenocysteine codons. PLoS Comput. Biol. 13: e1005367.
- Lin, H.C., Yeh, C.H., Chen, Y.F., Lee, T.T., Hsieh, P.Y., Rusnac, D.V., Lin, S.Y., Elledge, S.J., Zheng, N., Yen, H.C.S. (2018) C-terminal end-directed protein elimination by CRL2 ubiquitin ligases. Mol. Cell 70: 602- 613.
- Rusnac, D.V., Lin, S.C., Canzani, D., Tien, K., Hinds, T.R., Tsue, A.F., Bush, M.F., Yen, H.C.S., Zheng, N. (2018) Recognition of diglycine C-end degron by CRL2KLHDC2 ubiquitin ligase. Mol. Cell 72: 813-822.
- Yeh, C.W., Huang, W.C., Hsu, P.H., Yeh, K.H., Wang, L.C., Hsu, W.H., Lin, H.C., Chen, Y.N., Chen, S.C., Yeang, C.H., Yen, H.C.S. (2021) The C-degron pathway eliminates mislocalized proteins and products of deubiquitinating enzymes. EMBO J. 40: e105846.
- Hsu, Kuan-Lun, Yen, Hsueh-Chi S., and Yeang, Chen-Hsiang (2022). Cooperative stability renders protein complex formation more robust and controllable. Sci Rep 12, 10490.
- Yeh, C. W., Hsu, K. L., Lin, S. T., Huang, W. C., Yeh, K. H., Liu, C. J., Wang, L. C., Li, T. T., Chen, S. C., Yu, C. H., Leu, J. Y., Yeang, C. H., & Yen, H. S. (2024). Altered assembly paths mitigate interference among paralogous complexes. Nature communications, 15(1), 7169.