The Mechanism of RNA Decay
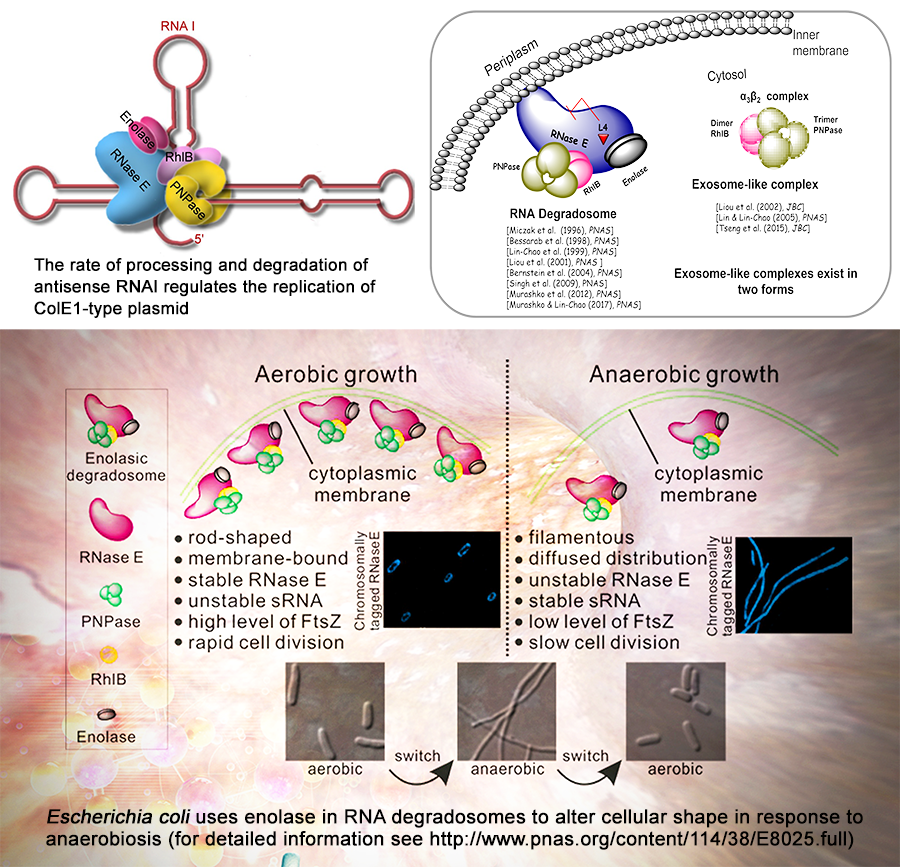
RNA 降解 RNA 降解對於細菌調控基因表現有重要的影響,而本研究室致力於探討 RNA 降解之分子機制。結合分子遺傳、分子生物、生物化學以及全基因組序列分析等技術,我們發現RNA 的降解速率可以調控 ColE1-型質體的複製以及細胞的生長; 我們純化並分析多核糖核酸水解酵素複合體(RNA 降解體;RNA Degradosome),發現 RNA 降解體是座落在細胞內膜,暗示細胞膜可能是RNA 降解體執行功能的場所,揭示此 RNA 降解體的完整性對於維持正常 RNA 代謝具有重要的角色,發現核醣體蛋白 L4 可以抑制此降解體的核酸內切活性,確定一個微型的“exosome-like RNA降解複合物”,揭示了烯醇化存在 RNA 降解體中的作用。 近期我們的研究工作則是致力於 RNA 降解在控制厭氧菌生長的重要及其分子機制。
細菌非編碼小 RNA 裁切處理和降解 細菌基因組包含許多非編碼小 RNA (non-coding small RNA)。最近的研究發現細菌利用 sRNA 調控細菌各種作為,包括防禦病毒感染和引發疾病。這些小的 RNA 多是由較大的原生型 RNA,經由核酸切割蛋白酵素(RNase)裁切處理而形成。 我們研究發現, RNase E 是裁切傳遞信使(tm- or ssrA-)原生RNA 的酵素,裁切形成的 tm-RNA,促使細菌具有正常的 tm-RNA 介導的肽蛋白標記功能,致使分解清除細菌內具肽標記的融合蛋白,維持蛋白質質量控制功能。 另外,我們發現細菌在厭氧下生長時,如何利用其 RNase E 降解體調控 DicF sRNA 的裁切處理,調控 DicF-sRNA 穩定與總量,致使其細胞性狀改變或存活。我們實驗室目前對細菌小 RNA 的研究,聚焦在研究、發現對細菌厭氧生長及生理功能有影響的 sRNA,並探討這些小 RNA 的生物功能和涉及的分子機制。
- PDF, 1987-1990, Dept. Genetics, Stanford Univ. Sch. Med., USA
- Ph.D., 1987, Dept. Mol. & Cell Biol., Univ. Texas-Dallas, USA
- MS, 1984, Dept. Mol. & Cell Biol., Univ. Texas-Dallas, USA
- BS, 1977, Dept. Biology, Natl. Changhua Normal Univ.
- Academia Sinica Investigator Award (2021)
- Taiwan Outstanding Women in Science Award (2020)
- Fellow Elected to the American Academy of Microbiology (2019)
- Merit MOST Research Fellow Award, Ministry of Science and Technology, Taiwan (2016)
- National Frontier Research Grant Award (1998-2003, 2007-2012)
- TWAS Prize in Biology, The World Academy of Sciences (2013)
- Academic Award, Ministry of Education, Taiwan (2009)
- Academia Sinica Young Investigator Award (1997)
- Outstanding Award, National Science Council/Ministry of Science and Technology, Taiwan (awarding continuously for 1995, 1997, 1999, 2002)
- The 33rd R.O.C. (Taiwan) Ten Outstanding Young Persons Award (1995)
RNA decay and bacterial cell growth
- Miczak, A, Kaberdin, V.R., Wei, C.-L., Lin-Chao, S. (1996) Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc Natl Acad Sci U S A. 93, 3865-3869.
- Kaberdin, V.R., Chao, Y.-H. and Lin-Chao, S. (1996) RNaseE cleaves at multiple sites in bubble regions of RNAI stem loops yielding products that dissociate differentially from the enzyme. J. Biol. Chem. 271, 13103-13109.
- Woo, W.-M. and Lin-Chao, S. (1997) Processing of the rne transcript by an RNase E-independent amino acid-dependent mechanism. J. Biol. Chem. 272, 15516-15520.
- Bessarab, D.A., Kaberdin, V.R., Wei, C.-L., Liou, G.-G. and Lin-Chao, S. (1998) RNA components of E. coli degradosome: Evidence for rRNA decay. Proc. Natl. Acad. Sci. USA 95, 3157-3161.
- Kaberdin, V.R., Miczak, A., Jakobsen, J.S., Lin-Chao, S., McDowall, K.J., and von Gabain, A. (1998) The endoribonucleolytic N-terminal half of Escherichia coli RNase E is evolutionarily conserved in Synechocystis sp. and other bacteria but not the C-terminal half, which is sufficient for degradosome assembly. Proc. Natl. Acad. Sci. U S A. 95, 11637-11642.
- Lin-Chao, S., Wei, C.-L., and Lin, Y.-T. (1999) RNase E is required for the maturation of ssrA RNA and normal ssrA RNA peptide-tagging activity. Proc. Natl. Acad. Sci. USA 96, 12406-12411.
- Liou, G.-G., Jane, W.-N., Cohen, S.N., Lin, N.-S., and Lin-Chao, S. (2001) RNA degradosomes exist in vivo in Escherichia coli as multi-component complexes associated with the cytoplasmic membrane via the N-terminal region of ribonuclease E. Proc. Natl. Acad. Sci. USA. 98:63-68.
- Bernstein, J. A., Khodursky, A. B., Lin, P.-H. , Lin-Chao, S. and Cohen, S. N.(2002) Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc. Natl. Acad. Sci. USA 99, 9697-9702.
- Liou, G.-G., Chang, H.-Y., Lin, C.-S., and Lin-Chao, S. (2002) DEAD box RhlB RNA helicase physically associates with exo-ribonuclease PNPase to degrade double-stranded RNA independent of the degradosome-assembling region of RNaseE. J Biol Chem. 277, 41157-41162.
- Bernstein, J.A., Lin, P.-H., Cohen, S.N. and Lin-Chao, S. (2004) Global analysis of Escherichia coli RNA degradosome function using DNA microarrays. Proc Natl Acad Sci U S A. 101, 2758-2763.
- Lin, P.-H., and Lin-Chao, S. (2005) RhlB helicase rather than enolase is the β-subunit of the Escherichia coli polynucleotide phosphorylase(PNPase)-exoribonucleolytic complex. Proc Natl Acad Sci U S A. 102, 16590-16595.
- Lin-Chao, S., Chiou, N.-T., and Schuster, G. (2007) The PNPase, exosome and RNA helicases as the building components of evolutionarily-conserved RNA degradation machines. J Biomed Sci. 14, 523-532.
- Lin, P.-H., Singh, D., Bernstein, J.A., and Lin-Chao, S. (2008) Genomic analysis of mRNA decay in E. coli with DNA microarrays. Methods Enzymol. 447, 47-64.
- Shi, Z., Yang, W.Z., Lin-Chao, S., Chak, K.F., and Yuan, H.S. (2008) Crystal structure of Escherichia coli PNPase: Central channel residues are involved in processive RNA degradation. RNA 14(11), 2361-71.
- Singh, D., Chang, S.-J., Lin, P.-H., Averina, O.V., Kaberdin, V.R., and Lin-Chao, S. (2009) Regulation of ribonuclease E activity by the L4 ribosomal protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 106, 864-869.
- Kaberdin VR, Lin-Chao S.(2009) Unraveling new roles for minor components of the E. coli RNA degradosome. RNA Biol. 2009 Sep-Oct;6(4):402-5.
- Kaberdin VR, Singh D, Lin-Chao S.(2011) Composition and conservation of the mRNA-degrading machinery in bacteria. J Biomed Sci. 2011 Mar 22;18:23
- Murashko, O.N., Kaberdin, V.R. and Lin-Chao, S. Membrane binding of Escherichia coli RNaseE catalytic domain stabilizes protein structure and increases RNA substrate affinity. Proc. Natl. Acad. Sci. U. S. A. 109 (18):7019-7024. 2012.
- Tseng YT, Chiou NT, Gogiraju R, *Lin-Chao S. The Protein Interaction of RNA Helicase B (RhlB) and Polynucleotide Phosphorylase (PNPase) Contributes to the Homeostatic Control of Cysteine in Escherichia coli. J Biol Chem. 290: 29953-29963, 2015
- Oleg N. Murashko and Lin-Chao, S.. Escherichia coli responds to environmental changes using enolasic degradosomes and stabilized DicF sRNA to alter cellular morphology PNAS 2017 doi: 10.1073/pnas.1703731114.
- Kaberdina AC, Ruiz-Larrabeiti O, Lin-Chao S and Kaberdin VR (2019) Correction to: Reprogramming of gene expression in Escherichia coli cultured on pyruvate versus glucose. Mol Genet Genomics. 2019 Oct;294(5):1373.
- Singh D, Murashko ON, Lin-Chao S.(2020) Post-transcriptional regulation of tnaA by protein-RNA interaction mediated by ribosomal protein L4 in Escherichia coli. J Bacteriol. 2020, 202(10):e00799-19
- Liou GG, Kaberdina AC, Wang WS, Kaberdin VR and Lin-Chao S (2022) Combined transcriptomic and proteomic profiling of E. coli under microaerobic versus aerobic conditions: the multifaceted roles of noncoding small RNAs and oxygen-dependent sensing in global gene expression control. Int. J. Mol. Sci. 2022, 23(5), 2570
- Murashko ON, Yeh KH, Yu CA, Kaberdin VR, Lin-Chao S (2023) Sodium fluoride exposure leads to ATP depletion and altered RNA decay in Escherichia coli under anaerobic conditions. Microbiol Spectr. 2023;11(2):e0415822.
- Wei-Syuan Wang and Sue Lin-Chao (2024) Hfq-Antisense RNA I Binding Regulates RNase E-Dependent RNA Stability and ColE1 Plasmid Copy Number. Int J Mol Sci. 2024 Apr 2;25(7):3955.
- Oleg N. Murashko, Connor Morgan-Lang, Chen-Hsin Albert Yu, Hsin-Nan Lin, Anna Chao Kaberdina, Shin-Yu Kung, Vladimir R. Kaberdin, Sue Lin-Chao (2025) Comprehensive analysis of repetitive extragenic palindrome sequences identified in bacteria and archaea using a new web-based tool, RepRanger. mSphere
- Wei-Syuan Wang, Yu-Hsiang Chen, Gunn-Guang Liou, Oleg N Murashko, Sue Lin-Chao (2025) The RNA degradation enzyme RNase E is essential for early flagellar assembly in Escherichia coli. PNAS Nexus
Mammalian growth arrest genes
- Lih, C.-J., Cohen, S.-N., Wang, C. and Lin-Chao, S. (1996) The platelet-derived growth factor alpha-receptor is encoded by a growth-arrest-specific (gas) gene. Proc Natl Acad Sci U S A. 93, 4617-4622.
- Ju, Y.-T., Chang, A.C.Y., She, B.-R., Tsaur, M.-L., Hwang, H.-M., Chao, C. C.-K., Cohen, S. N. and Lin-Chao, S. (1998) gas7: A gene expressed preferentially in growth-arrested fibroblasts and terminally differentiated Purkinje neurons affects neurite formation. Proc. Natl. Acad. Sci. USA 95, 11423-11428.
- Lazakovitch, E.M., She, B.-R., Lien, C.-L., Woo, W.-M., Ju, Y.-T., and Lin-Chao, S. (1999) The gas7 gene encodes two protein isoforms differentially expressed within the brain. Genomics 61, 298-306.
- She, B-R, Liou, G.-G., and Lin-Chao, S. (2002) Association of the growth arrest-specific protein Gas7 with F-actin induces reorganization of microfilaments and promotes membrane outgrowth Exp. Cell Res., 273: 34-44.
- Yeh, S.-D., Chen, Y.-J., Chang, A. C. Y., Ray, R., She, B. -R., Lee, W. -S., Chiang, H. -S., Cohen, S. N. and Lin-Chao, S. (2002) Isolation and properties of Gas8, a growth-arrest-specific gene regulated during male gametogenesis to produce a protein associated with the sperm motility apparatus. J. Biol. Chem., 277: 6311-6317.
- Chao, C. C.-K., Su, L.-J., Sun, N.-K., Ju, Y.-T., Lih, C.-J. and Lin-Chao, S. (2003) Involvement of Gas7 in NGF-independent and dependent cell processes in PC12 cells. J. Neuroscience Res. 74, 248-254.
- Chang, P.-Y., Kuo, J.-T., Lin-Chao, S. and Chao, C. C.-K. (2005) Identification of rat Gas7 isoforms differentially expressed in brain and regulated following kainate-induced neuronal injury. J. Neuroscience Res. 79, 788-797.
- Lortie, K., Huang, D., Chakravarthy, B., Comas, T., Hou, S.T., Lin-Chao, S., and Morley, P.(2005) The gas7 protein potentiates NGF-mediated differentiation of PC12 cells. Brain Res. 1036, 27-34.
- Chang, Y., Ueng, S.W.N., Lin-Chao, S., and Chao, C.C.-K. Involvement of Gas7 along the ERK 1/2 MAP kinase and SOX9 pathway in chondrogenesis of human marrow derived mesenchymal stem cells. Osteoarthritis Cartilage. 2008 Nov;16(11):1403-12.
- Su, W.B., You, J.-J., Huang, B.-T., Sivakumar, V., Yeh, S.-D., and Lin-Chao, S. Gas7. The Pombe Cdc15 Homology Proteins, Landes Bioscience, 2008
- You JJ, Lin-Chao S. (2010) Gas7 functions with N-WASP to regulate the neurite outgrowth of hippocampal neurons. J Biol Chem. Apr;285(15):11652-66.
- Wen, H.-L., Lin, Y.-T., Ting, C.-H., Lin-Chao, S., Li, H., Hsieh-Li, H.-M.(2010) Stathmin, a microtubule- destabilizing protein, is dysregulated in spinal muscular atrophy. Hum Mol Genet. 19(9) 1766-78.
- Hung, F.C., Chang, Y., Lin-Chao, S., and *Chao, C.C.K. Gas7 mediates the differentiation of human bone marrow-derived mesenchymal stem cells into functional osteoblasts by enhancing Runx2-dependent gene expression. J Orthop Res.10: 1528-35. 2011
- Huang, B.-T., Chang, P.-Y., Su, C.-H., Chao, C.C.K. and Lin-Chao, S.(2012) Gas7-deficient mouse reveals roles in motor function and muscle fiber composition during aging. PLoS ONE 7(5):e37702.(10.1371/journal.pone.0037702)
- Ting, C.-H., Wen, H.-L., Liu, H.-C., Hsieh-Li, H.-M., Li, H.and Lin-Chao, S.(2012) The Spinal Muscular Atrophy Disease Protein SMN Is Linked to the Golgi Network. PLoS ONE 7(12): e51826. doi:10.1371/journal.pone.0051826.
- Wen, H.-L., Ting, C.-H., Liu, H.-C., Li, H. and * Sue Lin-Chao.(2012) Decreased stathmin expression ameliorates neuromuscular defects but fails to prolong survival in a mouse model of spinal muscular atrophy. Neurobiology of Disease. 2013 Apr;52:94-103. doi: 10.1016/j.nbd.2012.11.015. Epub 2012 Dec 22.
- Liu, H.-C., Ting, C.-H., Wen, S.-L., Tsai, L.-K., Hsieh-Li, H.- M., Li, H. and *Lin-Chao S. Sodium vanadate combined with L-ascorbic acid delays disease progression, enhances motor performance, and ameliorates muscle atrophy and weakness in mice with spinal muscular atrophy. BMC Med. 2013 Feb 14;11(1):38. [Epub ahead of print]
- Tsai, L.-K., Chen, C.-L., Ting, C.-H., Lin-Chao, S, Hwu, W.-L., Dodge, J. C., Passini, M. and Cheng, S. Systemic administration of a recombinant AAV1 vector encoding IGF-1 improves disease manifestations in SMA mice. Mol Ther. 22(8):1450-9. doi: 10.1038/mt.2014.84. 2014
- Bhupana JN, Huang B-T, Liou G-G, Calkins MJ, Lin-Chao S. Gas7 knockout affects PINK1 expression and mitochondrial dynamics in mouse cortical neurons. FASEB BioAdvances.2020;00:1–16.