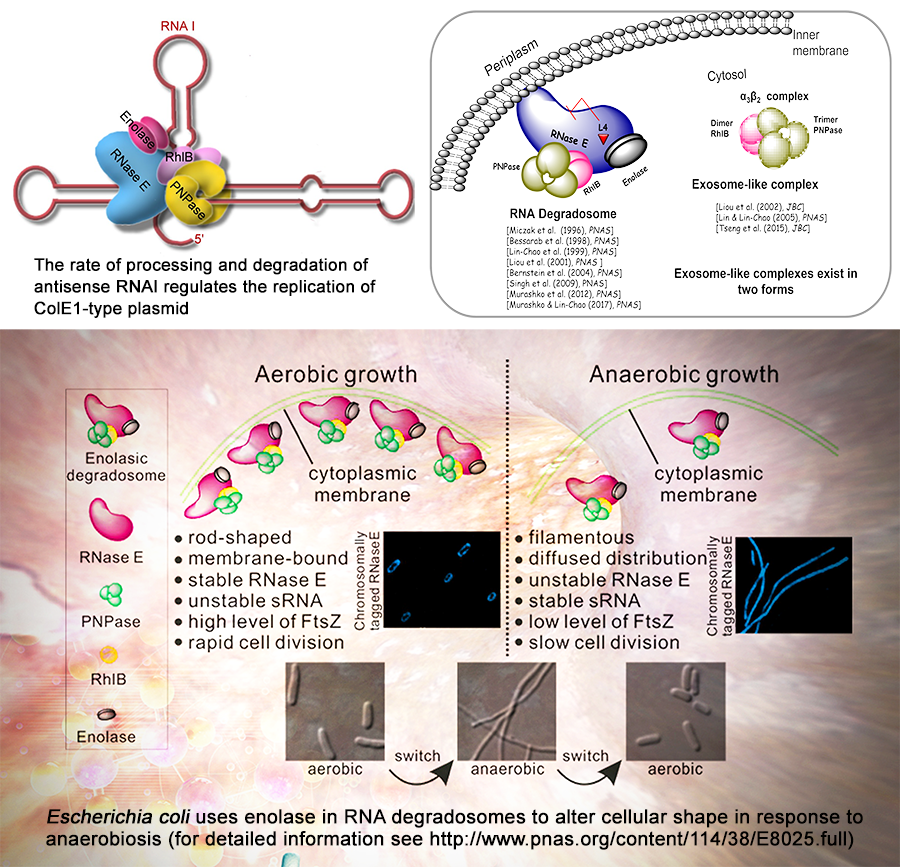
The Mechanism of RNA Decay
Our research interests focus on the mechanism of RNA decay and its important role in bacterial gene regulation during normal or adaptive growth. Using genetics, molecular biology, biochemical and genome-wide approaches, we have demonstrated the important roles of RNA decay in controlling DNA replication of ColE1-type plasmids and for normal cell growth. We have isolated and characterized RNA degradosomes (RNase E is scaffold protein, RhlB, PNPase and enolase are the three main protein components), characterized RNA degradosomes to be a cytoplasmic membrane-bound multicomponent ribonucleolytic complex in vivo, proved degradosome multi-enzymes work together to sustain normal RNA metabolism, showed ribosomal protein L4 to be an inhibitor of degradosome endonucleolytic functions, identified and characterized “exosome-like” mini- RNA degradation complexes, and uncovered the role of enolase in RNA degradosomes. Our current investigations are aimed at understanding the molecular mechanism by which RNA decay controls anaerobic cell growth.
Small RNA Processing and Stability
The bacterial genome contains many small RNAs. Recent studies have shown that small RNA is used to regulate various functions in bacteria, including defense against viral infections and pathogenesis. Most small RNAs are transcribed into larger precursors, which are further cleaved by endonuclease or exonuclease at the 5'end, 3'end, or both, and become functional RNA. We discovered that RNase E is required for the maturation of transfer-messenger (tm- or ssrA) RNA and normal tm-RNA mediated trans-translation peptide-tagging activity. We reported that the stability of DicF sRNA responds to oxygen levels through an enolase-RNase E degradosome-dependent mechanism, and controls anaerobic cell filamentation. Our current research on small RNAs aims to discover other sRNAs that have an effect on the anaerobic growth of bacteria and their physiological functions and to study the molecular mechanisms involved.
- PDF, 1987-1990, Dept. Genetics, Stanford Univ. Sch. Med., USA
- Ph.D., 1987, Dept. Mol. & Cell Biol., Univ. Texas-Dallas, USA
- MS, 1984, Dept. Mol. & Cell Biol., Univ. Texas-Dallas, USA
- BS, 1977, Dept. Biology, Natl. Changhua Normal Univ.
- Academia Sinica Investigator Award (2021)
- Taiwan Outstanding Women in Science Award (2020)
- Fellow Elected to the American Academy of Microbiology (2019)
- Merit MOST Research Fellow Award, Ministry of Science and Technology, Taiwan (2016)
- National Frontier Research Grant Award (1998-2003, 2007-2012)
- TWAS Prize in Biology, The World Academy of Sciences (2013)
- Academic Award, Ministry of Education, Taiwan (2009)
- Academia Sinica Young Investigator Award (1997)
- Outstanding Award, National Science Council/Ministry of Science and Technology, Taiwan (awarding continuously for 1995, 1997, 1999, 2002)
- The 33rd R.O.C. (Taiwan) Ten Outstanding Young Persons Award (1995)
RNA decay and bacterial cell growth
- Miczak, A, Kaberdin, V.R., Wei, C.-L., Lin-Chao, S. (1996) Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc Natl Acad Sci U S A. 93, 3865-3869.
- Kaberdin, V.R., Chao, Y.-H. and Lin-Chao, S. (1996) RNaseE cleaves at multiple sites in bubble regions of RNAI stem loops yielding products that dissociate differentially from the enzyme. J. Biol. Chem. 271, 13103-13109.
- Woo, W.-M. and Lin-Chao, S. (1997) Processing of the rne transcript by an RNase E-independent amino acid-dependent mechanism. J. Biol. Chem. 272, 15516-15520.
- Bessarab, D.A., Kaberdin, V.R., Wei, C.-L., Liou, G.-G. and Lin-Chao, S. (1998) RNA components of E. coli degradosome: Evidence for rRNA decay. Proc. Natl. Acad. Sci. USA 95, 3157-3161.
- Kaberdin, V.R., Miczak, A., Jakobsen, J.S., Lin-Chao, S., McDowall, K.J., and von Gabain, A. (1998) The endoribonucleolytic N-terminal half of Escherichia coli RNase E is evolutionarily conserved in Synechocystis sp. and other bacteria but not the C-terminal half, which is sufficient for degradosome assembly. Proc. Natl. Acad. Sci. U S A. 95, 11637-11642.
- Lin-Chao, S., Wei, C.-L., and Lin, Y.-T. (1999) RNase E is required for the maturation of ssrA RNA and normal ssrA RNA peptide-tagging activity. Proc. Natl. Acad. Sci. USA 96, 12406-12411.
- Liou, G.-G., Jane, W.-N., Cohen, S.N., Lin, N.-S., and Lin-Chao, S. (2001) RNA degradosomes exist in vivo in Escherichia coli as multi-component complexes associated with the cytoplasmic membrane via the N-terminal region of ribonuclease E. Proc. Natl. Acad. Sci. USA. 98:63-68.
- Bernstein, J. A., Khodursky, A. B., Lin, P.-H. , Lin-Chao, S. and Cohen, S. N.(2002) Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc. Natl. Acad. Sci. USA 99, 9697-9702.
- Liou, G.-G., Chang, H.-Y., Lin, C.-S., and Lin-Chao, S. (2002) DEAD box RhlB RNA helicase physically associates with exo-ribonuclease PNPase to degrade double-stranded RNA independent of the degradosome-assembling region of RNaseE. J Biol Chem. 277, 41157-41162.
- Bernstein, J.A., Lin, P.-H., Cohen, S.N. and Lin-Chao, S. (2004) Global analysis of Escherichia coli RNA degradosome function using DNA microarrays. Proc Natl Acad Sci U S A. 101, 2758-2763.
- Lin, P.-H., and Lin-Chao, S. (2005) RhlB helicase rather than enolase is the β-subunit of the Escherichia coli polynucleotide phosphorylase(PNPase)-exoribonucleolytic complex. Proc Natl Acad Sci U S A. 102, 16590-16595.
- Lin-Chao, S., Chiou, N.-T., and Schuster, G. (2007) The PNPase, exosome and RNA helicases as the building components of evolutionarily-conserved RNA degradation machines. J Biomed Sci. 14, 523-532.
- Lin, P.-H., Singh, D., Bernstein, J.A., and Lin-Chao, S. (2008) Genomic analysis of mRNA decay in E. coli with DNA microarrays. Methods Enzymol. 447, 47-64.
- Shi, Z., Yang, W.Z., Lin-Chao, S., Chak, K.F., and Yuan, H.S. (2008) Crystal structure of Escherichia coli PNPase: Central channel residues are involved in processive RNA degradation. RNA 14(11), 2361-71.
- Singh, D., Chang, S.-J., Lin, P.-H., Averina, O.V., Kaberdin, V.R., and Lin-Chao, S. (2009) Regulation of ribonuclease E activity by the L4 ribosomal protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 106, 864-869.
- Kaberdin VR, Lin-Chao S.(2009) Unraveling new roles for minor components of the E. coli RNA degradosome. RNA Biol. 2009 Sep-Oct;6(4):402-5.
- Kaberdin VR, Singh D, Lin-Chao S.(2011) Composition and conservation of the mRNA-degrading machinery in bacteria. J Biomed Sci. 2011 Mar 22;18:23
- Murashko, O.N., Kaberdin, V.R. and Lin-Chao, S. Membrane binding of Escherichia coli RNaseE catalytic domain stabilizes protein structure and increases RNA substrate affinity. Proc. Natl. Acad. Sci. U. S. A. 109 (18):7019-7024. 2012.
- Tseng YT, Chiou NT, Gogiraju R, *Lin-Chao S. The Protein Interaction of RNA Helicase B (RhlB) and Polynucleotide Phosphorylase (PNPase) Contributes to the Homeostatic Control of Cysteine in Escherichia coli. J Biol Chem. 290: 29953-29963, 2015
- Oleg N. Murashko and Lin-Chao, S.. Escherichia coli responds to environmental changes using enolasic degradosomes and stabilized DicF sRNA to alter cellular morphology PNAS 2017 doi: 10.1073/pnas.1703731114.
- Kaberdina AC, Ruiz-Larrabeiti O, Lin-Chao S and Kaberdin VR (2019) Correction to: Reprogramming of gene expression in Escherichia coli cultured on pyruvate versus glucose. Mol Genet Genomics. 2019 Oct;294(5):1373.
- Singh D, Murashko ON, Lin-Chao S.(2020) Post-transcriptional regulation of tnaA by protein-RNA interaction mediated by ribosomal protein L4 in Escherichia coli. J Bacteriol. 2020, 202(10):e00799-19
- Liou GG, Kaberdina AC, Wang WS, Kaberdin VR and Lin-Chao S (2022) Combined transcriptomic and proteomic profiling of E. coli under microaerobic versus aerobic conditions: the multifaceted roles of noncoding small RNAs and oxygen-dependent sensing in global gene expression control. Int. J. Mol. Sci. 2022, 23(5), 2570
- Murashko ON, Yeh KH, Yu CA, Kaberdin VR, Lin-Chao S (2023) Sodium fluoride exposure leads to ATP depletion and altered RNA decay in Escherichia coli under anaerobic conditions. Microbiol Spectr. 2023;11(2):e0415822.
- Wei-Syuan Wang and Sue Lin-Chao (2024) Hfq-Antisense RNA I Binding Regulates RNase E-Dependent RNA Stability and ColE1 Plasmid Copy Number. Int J Mol Sci. 2024 Apr 2;25(7):3955.
- Oleg N. Murashko, Connor Morgan-Lang, Chen-Hsin Albert Yu, Hsin-Nan Lin, Anna Chao Kaberdina, Shin-Yu Kung, Vladimir R. Kaberdin, Sue Lin-Chao (2025) Comprehensive analysis of repetitive extragenic palindrome sequences identified in bacteria and archaea using a new web-based tool, RepRanger. mSphere
- Wei-Syuan Wang, Yu-Hsiang Chen, Gunn-Guang Liou, Oleg N Murashko, Sue Lin-Chao (2025) The RNA degradation enzyme RNase E is essential for early flagellar assembly in Escherichia coli. PNAS Nexus
Mammalian growth arrest genes
- Lih, C.-J., Cohen, S.-N., Wang, C. and Lin-Chao, S. (1996) The platelet-derived growth factor alpha-receptor is encoded by a growth-arrest-specific (gas) gene. Proc Natl Acad Sci U S A. 93, 4617-4622.
- Ju, Y.-T., Chang, A.C.Y., She, B.-R., Tsaur, M.-L., Hwang, H.-M., Chao, C. C.-K., Cohen, S. N. and Lin-Chao, S. (1998) gas7: A gene expressed preferentially in growth-arrested fibroblasts and terminally differentiated Purkinje neurons affects neurite formation. Proc. Natl. Acad. Sci. USA 95, 11423-11428.
- Lazakovitch, E.M., She, B.-R., Lien, C.-L., Woo, W.-M., Ju, Y.-T., and Lin-Chao, S. (1999) The gas7 gene encodes two protein isoforms differentially expressed within the brain. Genomics 61, 298-306.
- She, B-R, Liou, G.-G., and Lin-Chao, S. (2002) Association of the growth arrest-specific protein Gas7 with F-actin induces reorganization of microfilaments and promotes membrane outgrowth Exp. Cell Res., 273: 34-44.
- Yeh, S.-D., Chen, Y.-J., Chang, A. C. Y., Ray, R., She, B. -R., Lee, W. -S., Chiang, H. -S., Cohen, S. N. and Lin-Chao, S. (2002) Isolation and properties of Gas8, a growth-arrest-specific gene regulated during male gametogenesis to produce a protein associated with the sperm motility apparatus. J. Biol. Chem., 277: 6311-6317.
- Chao, C. C.-K., Su, L.-J., Sun, N.-K., Ju, Y.-T., Lih, C.-J. and Lin-Chao, S. (2003) Involvement of Gas7 in NGF-independent and dependent cell processes in PC12 cells. J. Neuroscience Res. 74, 248-254.
- Chang, P.-Y., Kuo, J.-T., Lin-Chao, S. and Chao, C. C.-K. (2005) Identification of rat Gas7 isoforms differentially expressed in brain and regulated following kainate-induced neuronal injury. J. Neuroscience Res. 79, 788-797.
- Lortie, K., Huang, D., Chakravarthy, B., Comas, T., Hou, S.T., Lin-Chao, S., and Morley, P.(2005) The gas7 protein potentiates NGF-mediated differentiation of PC12 cells. Brain Res. 1036, 27-34.
- Chang, Y., Ueng, S.W.N., Lin-Chao, S., and Chao, C.C.-K. Involvement of Gas7 along the ERK 1/2 MAP kinase and SOX9 pathway in chondrogenesis of human marrow derived mesenchymal stem cells. Osteoarthritis Cartilage. 2008 Nov;16(11):1403-12.
- Su, W.B., You, J.-J., Huang, B.-T., Sivakumar, V., Yeh, S.-D., and Lin-Chao, S. Gas7. The Pombe Cdc15 Homology Proteins, Landes Bioscience, 2008
- You JJ, Lin-Chao S. (2010) Gas7 functions with N-WASP to regulate the neurite outgrowth of hippocampal neurons. J Biol Chem. Apr;285(15):11652-66.
- Wen, H.-L., Lin, Y.-T., Ting, C.-H., Lin-Chao, S., Li, H., Hsieh-Li, H.-M.(2010) Stathmin, a microtubule- destabilizing protein, is dysregulated in spinal muscular atrophy. Hum Mol Genet. 19(9) 1766-78.
- Hung, F.C., Chang, Y., Lin-Chao, S., and *Chao, C.C.K. Gas7 mediates the differentiation of human bone marrow-derived mesenchymal stem cells into functional osteoblasts by enhancing Runx2-dependent gene expression. J Orthop Res.10: 1528-35. 2011
- Huang, B.-T., Chang, P.-Y., Su, C.-H., Chao, C.C.K. and Lin-Chao, S.(2012) Gas7-deficient mouse reveals roles in motor function and muscle fiber composition during aging. PLoS ONE 7(5):e37702.(10.1371/journal.pone.0037702)
- Ting, C.-H., Wen, H.-L., Liu, H.-C., Hsieh-Li, H.-M., Li, H.and Lin-Chao, S.(2012) The Spinal Muscular Atrophy Disease Protein SMN Is Linked to the Golgi Network. PLoS ONE 7(12): e51826. doi:10.1371/journal.pone.0051826.
- Wen, H.-L., Ting, C.-H., Liu, H.-C., Li, H. and * Sue Lin-Chao.(2012) Decreased stathmin expression ameliorates neuromuscular defects but fails to prolong survival in a mouse model of spinal muscular atrophy. Neurobiology of Disease. 2013 Apr;52:94-103. doi: 10.1016/j.nbd.2012.11.015. Epub 2012 Dec 22.
- Liu, H.-C., Ting, C.-H., Wen, S.-L., Tsai, L.-K., Hsieh-Li, H.- M., Li, H. and *Lin-Chao S. Sodium vanadate combined with L-ascorbic acid delays disease progression, enhances motor performance, and ameliorates muscle atrophy and weakness in mice with spinal muscular atrophy. BMC Med. 2013 Feb 14;11(1):38. [Epub ahead of print]
- Tsai, L.-K., Chen, C.-L., Ting, C.-H., Lin-Chao, S, Hwu, W.-L., Dodge, J. C., Passini, M. and Cheng, S. Systemic administration of a recombinant AAV1 vector encoding IGF-1 improves disease manifestations in SMA mice. Mol Ther. 22(8):1450-9. doi: 10.1038/mt.2014.84. 2014
- Bhupana JN, Huang B-T, Liou G-G, Calkins MJ, Lin-Chao S. Gas7 knockout affects PINK1 expression and mitochondrial dynamics in mouse cortical neurons. FASEB BioAdvances.2020;00:1–16.