減數分裂
減數分裂是孟德爾遺傳學的核心。減數分裂是真核生物有性生殖細胞專有的細胞分裂方式,它將雙倍體的生殖細胞染色體數目減半,製造出單倍體的精子、卵或真菌有性孢子。 我們以麵包酵母(Saccharomyces cerevisiae)、粟酒裂殖酵母(Schizosaccharomyces pombe)與工業用瑞氏木黴菌(Trichoderma reesei)為研究模式,利用遺傳學、分子生物學、生物化學、結構生物學、基因體學與生物資訊學等方法,探討減數分裂細胞中同源重組(homologous recombination; HR)、 DNA 損傷反應(DNA damage response)、聯會複合體(synaptonemal complex; SC)與DNA 甲基轉移酶 (methyltransferase)如何產生遺傳變異與維持基因體完整性。
聯會複合體 是由DNA與蛋白質組成拉鍊狀的複雜結構,其核心功能是穩定地將同源染色體連結起來,讓姐妹染色分體能精確配對,為遺傳重組提供骨架,確保減數分裂成功和增加後代變異性的關鍵。聯會複合體於1956年被發現,但其真實的生化功能仍未知。
我們最近的研究成果與方向可分為三大項:
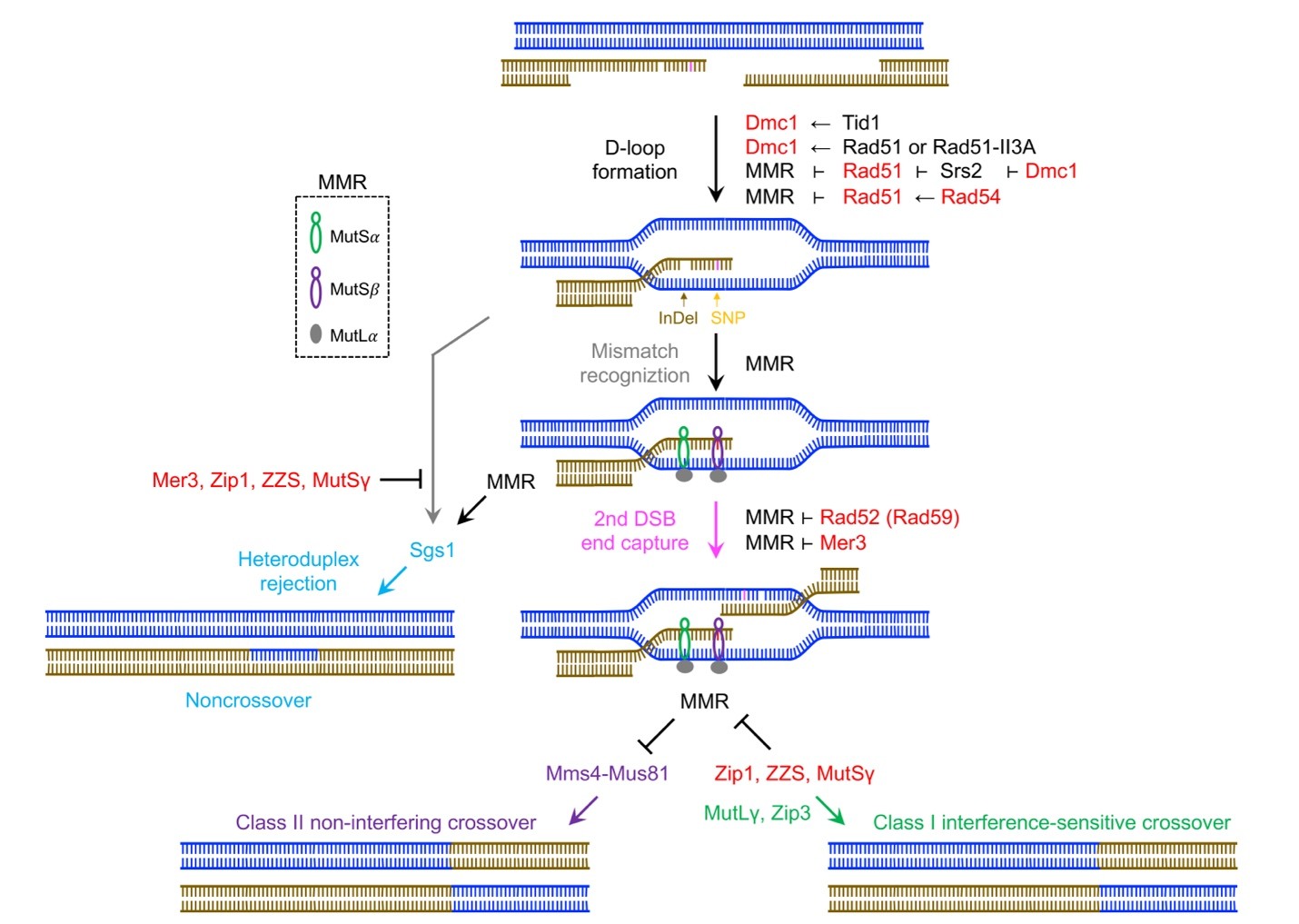
- 參與麵包酵母菌減數分裂的HR蛋白質(Rad51、Rad54與Rad59)和組成SC的ZMM蛋白質(Zip1、Zip4、 Mer3與Msh4)均能抑制「DNA錯誤配對鹼基修復(Mismatch repair; MMR)」的活性,是「同種異族」生物雜交繁衍不可或缺的要件。我們利用全基因體研究數據與其它相關文獻資料,解釋過去30年來生物學與遺傳學教科書中關於「MMR導致相近物種雜交不孕與生殖隔離」舊理論的謬誤,再以「反向遺傳學(reverse genetics)」研究方法,找到12個專一性影響「同種異族雜交」酵母菌減數分裂的HR突變基因,相同的突變基因卻不影響「純品系」酵母菌減數分裂,證明多個HR蛋白質能抑制MMR活性,保留錯誤配對DNA鹼基。這個新發現的重要性之一是挑戰「HR是修補破損DNA最高保真度的機制」的舊學說。聯會複合體發現迄今近70年,但真正的生理功能仍未知。我們首次證明聯會複合體在減數分裂負責抑制MMR,促進DNA序列不完全相同的同源染色體進行重組互換,增加配子或真菌有性孢子的遺傳多樣性。
- 大多數進行有性生殖的真核生物(如酵母菌、擔子菌、果蠅、線蟲、高等植物與哺乳類)的減數分裂細胞,都利用 Spo11 蛋白產生 DNA 雙股斷裂,從而啟動HR反應。我們發現剔除 spo11 基因的木黴菌仍能正常進行減數分裂與產生有新遺傳變異的有性孢子。 目前研究重點在探討木黴菌如何能在沒有Spo11啟動減數分裂HR反應。
- 我們發現瑞氏木黴菌的DNA甲基轉移酶在減數分裂中具多項功能,第一是負責啟動減數分裂,第二是促進親代雙方同源染色體間HR反應,產生新遺傳變異。第三是以特殊表觀遺傳機制,將親代DNA上大量的胞嘧啶(cytosine) 甲基化,從而誘發點突變成胸腺嘧啶(thymine),快速增加減數分裂產物(有性孢子)的基因體多樣性。遺傳變異過高的效應有如雙面刃,雖然有時能提升後代環境適應能力,但也常導致基因體產生不穩定性,造成有性孢子無法萌發成菌絲,又或者萌發出的新生菌絲無法正常進行營養生長。此新理論能解釋為何多數的野生木黴菌均為雌性或雄性不孕,僅憑藉菌絲營養生長而無性繁衍,因為更能維持基因體穩定性並延續既有生存優勢。
不同物種生物雜交後,生殖細胞進行減數分裂,親代雙方同源染色體各自複製成一對姊妹染色質,然後DNA進行同源性重組(homologous recombination; HR)反應,促進同源染色體間遺傳物質互換,增加精子、卵與真菌有性孢子的基因多樣性。我們發現參與促進HR反應的不同蛋白質,在多個HR反應關鍵步驟抑制「錯誤配對鹼基修復(mismatch repair; MMR)」,例如「形成D環」、「捕捉第二個DNA雙股斷裂」與「形成ZMM蛋白質依賴性或第一類交叉產物(crossover; CO)」。MMR抑制非ZMM蛋白質依賴性或第二類CO產物。以抗老化知名的DNA解旋酶Sgs1可以MMR依賴性與MMR非依賴性兩種方式,促進非交叉產物(noncrossover; CO)形成。紅色標記所有具有「抗MMR活性」的HR蛋白質。親代雙方同源染色體DNA進行HR反應,因DNA序列不完全相同,形成具錯誤配對鹼基的異源雙鏈體(heteroduplex DNA),會被三個MMR蛋白複合體(MutSα, MutSβ, and MutLα)辨識與修補,從而阻止雜交減數分裂HR正常進行。催化HR反應的兩個關鍵酵素Rad51與Dmc1,分別利用不同的機制耐受錯誤配對鹼基與抑制MMR作用。Dmc1本身與其輔助蛋白質Tid1均不具抑制MMR活性,但較Rad51能耐受錯誤配對鹼基。具抗Rad51活性的DNA解旋酶Srs2,負責分解Rad51與破損的單股DNA形成的核蛋白纖維絲複合體,因此可被歸類成一個新型「MMR蛋白質」。Dmc1能抑制Srs2解旋酶活性,負負得正,Dmc1因此也成為「抗MMR活性」蛋白質。
- PDF, 1999-2000, Dept. Mol. & Cell Biol., Harvard Univ. USA
- Ph.D., 1998, Dept. Mol. & Cell Biol., Harvard Univ. USA
- MS, 1990, Inst. Life Science., Natl. Tsing Hua Univ.
- BS, 1988, Dept. Chemistry, Natl. Taiwan Univ.
- Ting-Fang Wang# and Ji-Long Liao. (2025) Homologous recombination counteracts mismatch repair to promote fertility and genetic diversity. Trends in Genetics (PMID: 41241584; DOI: 10.1016/j.tig.2025.10.009)
- Ya-Ling Hung, Chi-Ning Chuang, Hong-Xiang Kim, Hou-Cheng Liu, Jhong-Syuan Yao, Lavernchy Jovanska, Yi-Ping Hsueh, Ruey-Shyang Chen, and Ting-Fang Wang# (2025) Rad51, Rad54, and ZMM proteins antagonize the mismatch repair system to promote fertility of budding yeast intraspecies hybrid zygotes. Nucleic Acids Research (PMID: 40902007; DOI: 10.1093/nar/gkaf847)
- Hsiao-Tang Hu, Ueh-Ting Tim Wang, Bi-Chang Chen, Yi-Ping Hsueh#, Ting-Fang Wang# (2025) Ki-67 and CDK1 control the dynamic association of nuclear lipids with mitotic chromosomes. Journal of Lipid Research ( PMID: 39706365; DOI: 10.1016/j.jlr.2024.100731)
- Lavernchy Jovanska, I-Chen Lin, Jhong-Syuan Yao, Chia-Ling Chen, Hou-Cheng Liu, Wan-Chen Li, Yu-Chien Chuang, Chi-Ning Chuang, Albert Chen-Hsin Yu, Hsin-Nan Lin, Wen-Li Pong, Chang-I Yu, Ching-Yuan Su, Yi-Ping Chen, Ruey-Shyang Chen, Yi-Ping Hsueh, Hanna S Yuan, Ljudmilla Timofejeva, Ting-Fang Wang# (2024) DNA cytosine methyltransferases differentially regulate genome-wide hypermutation and interhomolog recombination in Trichoderma reesei meiosis. Nucleic Acids Research (PMID: 39021337; DOI: 10.1093/nar/gkae611)
- Chi-Ning Chuang, Hou-Cheng Liu, Tai-Ting Woo, Ju-Lan Chao, Chiung-Ya Chen, Hisao-Tang Hu, Yi-Ping Hsueh, Ting-Fang Wang# (2024) Noncanonical usage of stop codons in ciliates expands proteins with structurally flexible Q-rich motifs. eLife (PMID: 38393970; DOI: 10.7554/eLife.91405)
- Chia-Lin Chen, Wan-Chen Li, Yu-Chien Chuang, Hou-Cheng Liu, Chien-Hao Huang, Ko-Yun Lo, Chung-Yu Chen, Fang-Mo Chang, Guo-An Chang, Yu-Ling Lin, Wen-denr Yang, Ching-Hua Su, Tsung-Ming Yeh, and Ting-Fang Wang# (2022) Sexual crossing, chromosome-level genome sequences, and comparative genomic analyses for the medicinal mushroom Taiwanofungus camphoratus (Syn. Antrodia cinnamomea; Antrodia camphorata). Microbiology spectrum (PMID: 35196809; DOI: 10.1128/spectrum.02032-21)
- Wan-Chen Li, Ting-Chan Lin, Chia-Ling Chen, Hou-Cheng Liu, Hisn-Nan Lin, Ju-Lan Chao, Cheng-Hsilin Hsieh, Hui-Fang Ni, Ruey-Shyang Chen# and Ting-Fang Wang# (2021) Complete genome sequences and genome-wide characterization of Trichoderma biocontrol agents provide new insights into their evolution and variation in genome organization, sexual development, and fungal-plant interactions. Microbiology spectrum (PMID: 34908505; DOI: 10.1128/Spectrum.00663-21)
- Wan-Chen Li, Chia-Yi Lee, Wei-Hsuan Lan, Tai-Ting Woo, Hou-Cheng Liu, Hsin-Yi Yeh, Hao-Yan Chang, Yu-Chien Chuang, Chi-Ning Chuang, Chia-Lin Chen, Yi-Ping Hsueh, Hung-Wen Li#, Peter Chi#, and Ting-Fang Wang# (2021) Trichoderma reesei Rad51 tolerates mismatches in hybrid meiosis with diverse genome sequences. Proceedings of the National Academy of Sciences (PMID: 33593897; DOI: 10.1073/pnas.2007192118)
- Wan-Chen Li and Ting-Fang Wang# (2021) PacBio long-read sequencing, assembly, and Funannotate reannotation of the complete genome of Trichoderma reesei QM6a. Methods in Molecular Biology (PMID: 33165795; DOI: 10.1007/978-1-0716-1048-0_21)
- Hou-Cheng Liu, Wan-Chen Li, and Ting-Fang Wang# (2021) TSETA: A third-generation sequencing-based computational tool for mapping and visualization of SNPs, meiotic recombination products, and RIP Mutations. Methods in Molecular Biology (PMID: 33165796; DOI: 10.1007/978-1-0716-1048-0_2)
- Tai-Ting Woo, Chi-Ning Chuang, Mika Higashide, Akira Shinohara, and Ting-Fang Wang# (2020) Dual roles of yeast Rad51 N-terminal domain in repairing DNA double-strand breaks. Nucleic Acids Research (PMID: 32652040; DOI: 10.1093/nar/gkaa587)
- Wan-Chen Li, Hou-Cheng Liu, Ying-Jyun Lin, Shu-Yun Tung, Ting-Fang Wang# (2020) Third-generation sequencing-based mapping and visualization of single nucleotide polymorphism, meiotic recombination, illegitimate mutation and repeat-induced point mutation. NAR Genomics and Bioinformatics (PMID: 33575607; DOI: 10.1093/nargab/lqaa056)
- Wang, T.-F., Chin-Ning Chuang and Tai-Ting Woo. “Method and vector for enhancing protein expression”. US provisional application (63/036,704), Patent Cooperation Treaty application (PCT/US2021/036247) and Taiwan application (110120541).
- Wang, T.-F., “Method for producing segmental aneuploidy (SAN) strains of Trichoderma reesei via sexual crossing and SAN strains produced therefrom”. Patent Cooperation Treaty (PCT/US14/58654) and US (9598738B2), Taiwan (I570236) and People’s Republic of China (IA001/ACA081CN).
- Wang, T. F. and Yang, S.M. “Methods of producing virus-like particles of piconavirus using a small-ubiquitin-related fusion protein expression system”. US (8663951B2) and Taiwan (I415623).
- Wang, T.-F. “SUMO fusion protein expression system for producing native proteins”. USA 8034910BZ.