RNA Splicing in Disease
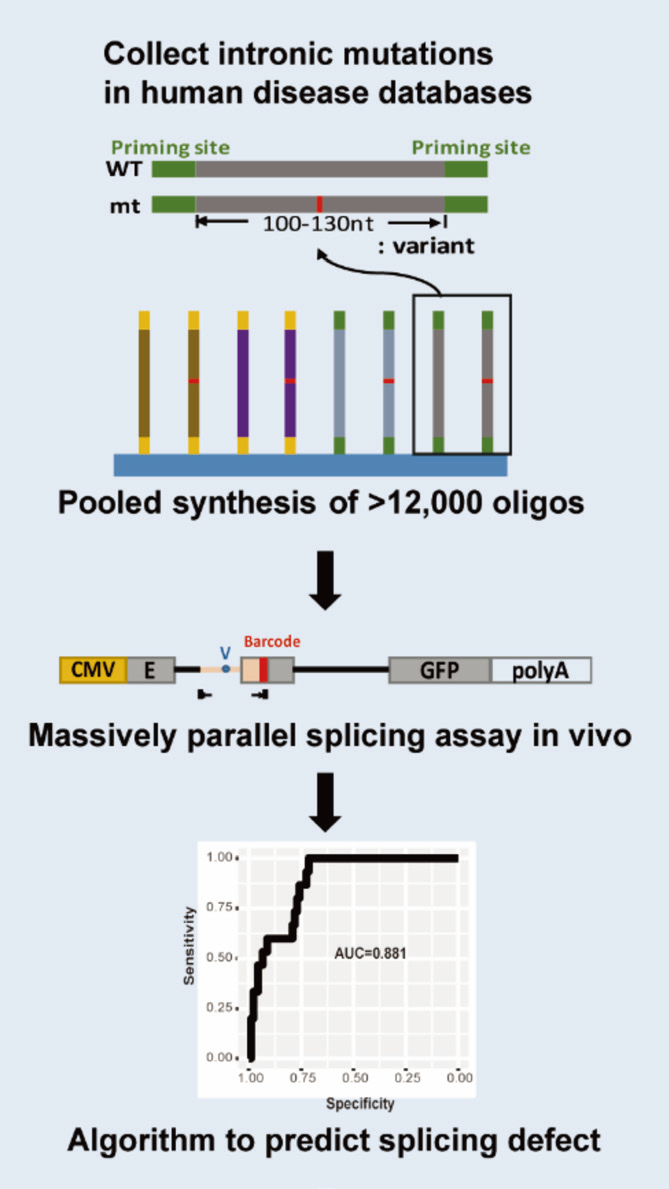
Post-transcriptional control is the most important regulation to maximize the genome capacity and to fine-tune the gene expression. After transcribed from DNA, the intervening non-coding sequence, introns, will be removed, and exons, will be ligated. This process is pre-mRNA splicing. Pre-mRNA splicing is a highly regulated process with remarkable precision. The estimation is that ~10% genetic variants cause diseases through RNA splicing defects. However, with such high importance, the mechanism of the splicing regulation is not fully understood. In our lab, with interdisciplinary approaches, we study how splice sites are recognized and how splicing responds to genetic variants in a large scale. We developed a highthroughput splicing assay that enables multiplexed examination of thousands of disease-relevant genetic variants simultaneously. We combined molecular biology, genomics, computational biology and machine learning to dissect the splicing decisions and are able to predict the splicing defect of novel genetic variants. We aim to decipher the splicing regulation of the natural genetic variants in the normal and the disease background for the promise of precision medicine.
RNA Stability and Translation Control in Disease
Following splicing, the mature mRNA is transported to subcellular compartments and awaits proper signals for translation. The untranslated regions (UTRs) of mRNA contain many sequence elements that are critical for its stability and translation regulation. To systematically investigate if the RNA stability and translation level is affected by the disease-related mutations, we use massively parallel reporter assays to estimate the stability of each UTR over time course and their translation efficiency by their distribution in the polyribosome profile. We found that the primary sequence as well as the secondary structure are equally critical in determining the RNA destiny. This study aims to reveal the contribution of the noncoding region in controlling gene expression in the normal and the disease background.
- PDF, 2012-2016, Dept. Mol. Cell. Bio. & Biochem., Brown Univ., USA
- Ph.D., 2005-2012, Mol. Medicine, Univ. Mass. Medical School, USA
- MS, 2002-2003, Neuroscience, Univ. College London, UK
- BS, 1998-2002, Dept. Zoology, Natl.Taiwan Univ. Taiwan
- 2017-2019, Initial Employment Academic Research Grants 中研院新聘學術獎, Academia Sinica, Taiwan
- 2017-2020, MOST Talented Scholar Fellowship 科技部延攬特殊優秀人才補助, Ministry of Science and Technology, Taiwan
- 2020-2023, Career Development Award, National Health Research Institutes, Taiwan
- 2020-2022, Outstanding Young Scholar Research Grant , Ministry of Science and Technology, Taiwan
- 2020-2022, The Young Scholars' Creativity Award 傑出人才基金會年輕學者創新獎, The Foundation for the Advancement of Outstanding Scholarship, Taiwan
- 2012-current, member, RNA Society
- 2023, EMBO Global Investigator 歐洲分子生物學組織「全球研究學者」
- Lin, C.-L.*, Taggart, A. J.*, Lim, K. H.*, Cygan, K. J., Ferraris, L., Creton, R., Huang, Y.-T., Fairbrother, W. G. (2016) RNA structure replaces the need for U2AF2 in splicing. Genome Research 26: 12-23. *equal contribution
- Lin, C.-L., Taggart, A. J., Fairbrother, W. G. (2016) RNA structure in splicing: an evolutionary perspective. RNA Biol. 13: 766-771.
- Taggart, A. J.*, Lin, C.-L.*, Shrestha, B., Heintzelman, C., Kim, S. W., Fairbrother, W. G. (2017) Large-scale analysis of branchpoint usage across species and cell lines. Genome Research 27: 639-649. *equal contribution
- Chiang, H.-L.*, Chen, Y.-T.*, Su, J.-Y.*, Lin, H.-N., Yu, A. C.-H., Hung, Y.-J., Wang, Y.-L., Huang, Y.-T., Lin, C.-L. (2022) Mechanism and modeling of human disease-associated near-exon intronic variants that perturb RNA splicing. Nature Structural & Molecular Biology 29: 1043–1055. *equal contribution
- Huang, A.-C.*, Chiang H.-L.*, Hung Y.-J.*, Chen, Y.-T., Su, J.-Y., Huang, Y-.T., Yu, A. C.-H., Lin, H.-N.#, Lin, C.-L.# (2024) SpliceAPP: An Interactive Web Server to Predict Splicing Errors Arising from Human Mutations. BMC Genomics 25:600. *equal contribution #corresponding authors
- Su, J.-Y.*, Wang, Y.-L.*, Chang, Y-.C., Hsieh Y.-T., Yang, C.-H., Kang, Y., Huang, Y.-T., Lin, C.-L. (2025) Multiplexed Assays of Human Disease-relevant Mutations Reveal UTR Dinucleotide Composition as a Major Determinant of RNA Stability. eLife 13:RP97682. *equal contribution

