Exploring Chromatin Biology through Functional Genomics
Chromatin regulators, including histones, modifying enzymes, transcription factors, and chromatin remodelers, form the fundamental machinery that controls access to the genome. Notably, genes encoding these factors represent one of the most frequently mutated gene classes in cancer. We study how chromatin-associated factors regulate gene expression and how their disruption contributes to cancer. Using functional genomics approaches, our lab seeks to uncover how altered chromatin regulation drives disease.
Our research group investigates how disease-associated mutations in chromatin regulators disrupt gene regulation.
We integrate functional genomics and mechanistic approaches to trace effects of genetic variation from sequence → mechanism → phenotype , with the goal of identifying actionable cancer vulnerabilities.
We pursue this through a systematic framework:
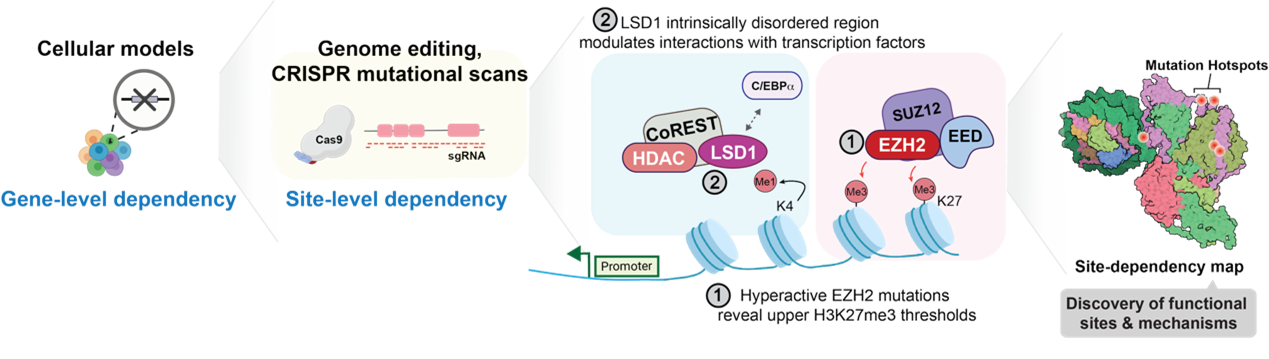
- Discovery: Identify mutations in chromatin regulators that have functional and cellular impact
- Mechanism: Define how prioritized mutations alter chromatin function at the molecular level.
- Translation: Connect molecular mechanisms to cellular phenotypes and therapeutic opportunities.
A central focus of the lab is the study of recurrent mutations in lysine methyltransferase complexes and histone proteins in cancer.
- PDF, 2019-2025, Department of Chemistry and Chemical Biology, Harvard University
- PDF, 2017-2019, Department of Molecular Biophysics and Biochemistry, Yale University
- Ph.D., 2011-2016, NUS Graduate School – Integrative Sciences and Engineering Program, National University of Singapore (NUS)
- B.S., 2007-2010, School of Biological Sciences, Nanyang Technological University, Singapore
- 2025, Forbeck Foundation Scholar Award
- 2023, Abstract Achievement Award, American Society of Hematology (ASH).
- 2023, Eliana Hechter Memorial Travel Prize, Broad Institute.
- 2022, Charles A. King Trust Postdoctoral Research Fellowship, Health Resources in Action.
- Kwok HS*, Morriss JW*, Hu CXY*, Iram I, Myung Y, Siegenfeld AP, Waterbury AL, Yeo MJR, Lee C, Camp S, Iqbal S, Liau BB. (2025) Base editing charts a sequence-function atlas of Polycomb Repressive Complex 2 at amino acid resolution. Submitted.
- Kwok HS*, Freedy AM*, Siegenfeld AP, Morriss JW, Waterbury AL, Kissler SM, Liau BB. (2023) Drug addiction mutations unveil a methylation ceiling in EZH2-mutant lymphoma. Nature Chemical Biology. 19, 1105-1115. doi: 10.1038/s41589-023-01299-1.
- Waterbury AL*, Kwok HS*, Lee C*, Narducci D, Freedy AM, Su C, Raval S, Reiter A, Hawkins W, Lee K, Li J, Hoenig SM, Vinyard ME, Cole PA, Hansen AS, Carr SA, Papanastasiou M, Liau BB. (2024) An autoinhibitory switch of the LSD1 disordered region controls enhancer silencing. Molecular Cell, 84, 2238-2254. doi: 10.1016/j.molcel.2024.05.017
- Yeo MJR*, Zhang O*, Xie X*, Nam E*, Payne NC, Gosavi PM, Kwok HS, Iram I, Lee C, Li J, Chen N, Nguyen K, Jiang H, Wang ZA, Lee K, Mao H, Harry SA, Barakat I, Takahashi M, Waterbury AL, Barone M, Mattevi A, Carr SA, Udeshi ND, Bar-Peled L, Cole PA, Mazitschek R, Liau BB.#, Zheng N.# (2025) UM171 glues asymmetric CRL3–HDAC1/2 assembly to degrade CoREST corepressors. Nature, 639, 232–240. doi.org/10.1038/s41586-024-08532-4 (#Co-corresponding authors)
- Xie X*, Zhang O*, Yeo MJR*, Lee C*, Tao R, Harry SA, Payne NC, Nam E, Paul L, Li Y, Kwok HS, Jiang H, Mao H, Hadley JL, Lin H, Batts M, Gosavi PM, D’Angiolella V, Cole PA, Mazitschek R, Northcott PA, Zheng N#, Liau BB#. (2025) Converging mechanism of UM171 and KBTBD4 neomorphic cancer mutations. Nature, 639, 241–249. doi.org/10.1038/s41586-024-08533-3 (#Co-corresponding authors)
- Kwok HS#, Vargas-Rodriguez O, Melnikov SV, Söll D. (2019) Engineered aminoacyl-tRNA synthetases with improved selectivity toward noncanonical amino acids. ACS Chemical Biology, 14(4), 603–612. doi: 10.1021/acschembio.9b00088 (# Sole corresponding author)
- Numata A*, Kwok HS*, Zhou QL*, Li J, Tirado-Magallanes R, Angarica VE, Hannah R, Park J, Wang CQ, Krishnan, V, Rajagopalan D, Zhang Y, Zhou S, Welner RS, Osato M, Jha S, Bohlander SK, Göttgens B, Yang H, Benoukraf T, Lough JW, Bararia D, Tenen DG. (2020) Lysine acetyltransferase Tip60 is required for hematopoietic stem cell maintenance. Blood, 136(15), 1735–1747. doi: 10.1182/blood.2019001279
- Numata A*, Kwok HS*, Kawasaki A, Li J, Zhou QL, Kerry J, Benoukraf T, Bararia D, Li F, Ballabio E, Tapia M, Deshpande AJ, Welner RS, Delwel R, Yang H, Milne TA, Taneja R, Tenen DG. (2018) The basic helix-loop-helix transcription factor SHARP1 is an oncogenic driver in MLL-AF6 acute myelogenous leukemia. Nature Communications, 9(1), 1622. doi: 10.1038/s41467-018-03854-0
- Bararia D*, Kwok HS*, Welner RS, Numata A, Sárosi MB, Yang H, Wee S, Tschuri S, Ray D, Weigert O, Levantini E, Ebralidze AK, Gunaratne J, Tenen DG. (2016) Acetylation of C/EBPα inhibits its granulopoietic function. Nature Communications, 7(1), 10968. doi: 10.1038/ncomms1096
* denotes co-first authors, # denotes corresponding authors