Membrane fluidity, lipid homeostasis, proteostasis, organelle dynamics
Lipids serve as the structural component of cellular membranes, energy sources and signaling molecules. Fluid membranes allows for dynamic events, including membrane fusion and fission, endocytosis and exocytosis, organelle segregation during cell division, and protein movement within the membrane. Membrane fluidity is largely determined by the level of unsaturated lipids and sterols that govern the order of lipid packing in the membrane. Unsaturated fatty acids, with double bonds in their carbohydrate chains, are essential for life. It remains largely unclear how lipid saturation impacts the function and dynamics of membranes at the cellular level. To address this question, we resort to the budding yeast Saccharomyces cerevisiae which is advantageous over other systems as it contains single fatty acid desaturase gene OLE1. We have generated temperature-sensitive ole1 mutants to investigate the cellular function of unsaturated fatty acids, using genetic, biochemical, cell biological, and systematic analysis. Our current focus includes the following:
- The impact of lipid saturation on the overall membrane architecture and dynamics.
- The coordinated regulation of unsaturated fatty acids and sterol biosynthesis for the maintenance of membrane homeostasis.
- The impact of lipid saturation on membrane protein folding and the cellular response to this stress.
Our goal is to eventually understand at molecular details how the perturbation of lipid saturation is sensed and relayed to the control of lipid homeostasis and proteostasis and how the process impacts cellular physiology.
- PDF, 1994-1997, University of California, San Francisco USA
- Ph.D., 1993, Department of Cell. & Mol. Physiology Harvard University, USA
- D.D.S., 1987, Department of Dentistry National Taiwan University
- 2005-2009, Academia Sinica Investigator Award
- 1998-2003, David and Lucile Packard Award in Science and Engineering, USA
- 1994-1997, Helen Hay Whitney Postdoctoral Research Fellowship, USA
- Cheng, Y.-L., Kuan, J.-E., Wang, C.-W., Chen, R.-H. 2025. Mga2-mediated transcription supports mitotic nuclear expansion under lipid saturation conditions in stearoyl-CoA desaturase Ole1 mutant. Mol. Biol. Cell 36(11):ar144, 1-17.
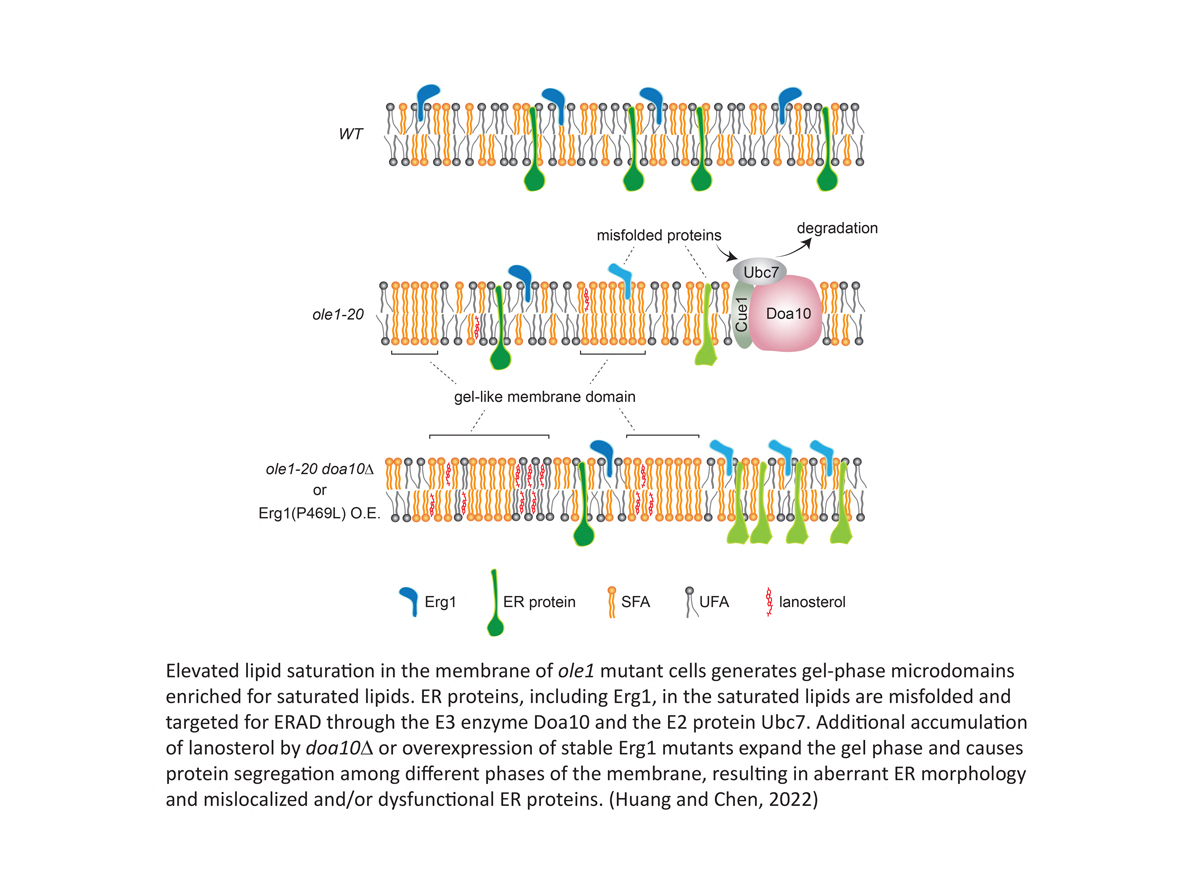
- Huang, L.-J. and Chen, R.-H. 2022. Lipid saturation induces degradation of squalene epoxidase for sterol homeostasis and cell survival. Life Sci. Alliance 6(1):e202201612.
- Chen, R.-H. 2019. Chromosome detachment from the nuclear envelope is required for genome stability in closed mitosis. Mol. Biol. Cell 30(13):1578-1586.
- Hsu, T.-H., Chen, R.-H., Cheng, Y.-H., and Wang, C.-W. 2017. Lipid droplets are central organelles for meiosis II progression during yeast sporulation. Mol. Biol. Cell 28(3): 440-451.
- Yang, P.-L., Hsu, T.-H., Wang, C.-W., and Chen, R.-H. 2016. Lipid droplets maintain lipid homeostasis during anaphase for efficient cell separation in budding yeast. Mol. Biol. Cell 27(15):2368-2380.
- Cheng, Y.-L. and Chen, R.-H. 2015. Assembly and quality control of the protein phosphatase 1 holoenzyme involves the Cdc48-Shp1 chaperone. J. Cell Sci. 128(6):1180-1192.
- Madsen, L., Molbæk, K., Larsen, I.B., Nielsen, S.V., Poulsen, E.G., Walmod, P.S., Hofmann, K., Seeger, M., Chien, C.Y., Chen, R.-H., Kriegenburg, F., Hartmann-Petersen, R. 2014. Human ASPL/TUG interacts with p97 and complements the proteasome mislocalization of a yeast ubx4 mutant, but not the ER-associated degradation defect. BMC Cell Biol. 15:31.
- Chien, C.-Y. and Chen, R.-H. 2013. Cdc48 chaperone and adaptor Ubx4 distribute the proteasome in the nucleus for anaphase proteolysis. J. Biol. Chem. 288(52): 37180-37191.
- Chen, R.-H. 2012. Nuclear envelope assembly and disassembly during the cell cycle. eLS. John Wiley & Sons, Ltd: Chichester.
- Tseng, L.-C. and Chen, R.-H. 2011. Temporal control of nuclear envelope assembly by phosphorylation of lamin B receptor. Mol. Biol. Cell 22:3306-3317.
- Hsieh, M.-T. and Chen, R.-H. 2011. Cdc48 and cofactors Npl4-Ufd1 are important for G1 progression during heat stress by maintaining cell wall integrity in Saccharomyces cerevisiae. PLoS ONE 6: e18988.
- Cheng, Y.-L. and Chen, R.-H. 2010. AAA-ATPase Cdc48 and Cofactor Shp1 promote chromosome bi-orientation by balancing Aurora B activity. J. Cell. Sci. 123: 2025-2034.