SpliceAPP: An Interactive Web Server to Predict Splicing Errors Arising from Human Mutations
Dr. Lin, Chien-Ling - July, 2024
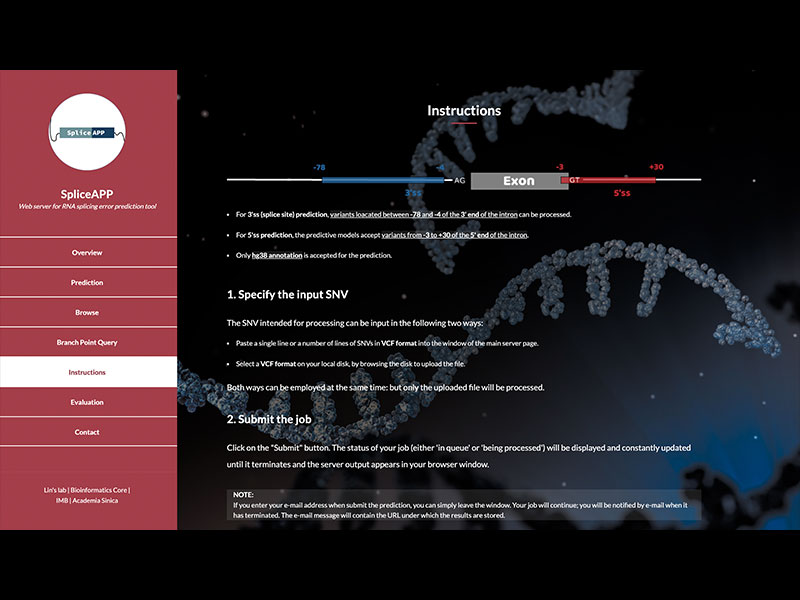
Splicing variants are a major class of pathogenic mutations, but redundant and degenerate splicing signals hinder functional assessments of sequence variations within introns. To facilitate the functional annotation of splicing variants, Dr. Chien-Ling Lin's group and the IMB Bioinformatics Core, led by Drs. Hsin-Nan (Arith) Lin and Chen-Hsin (Albert) Yu, developed a web-based tool, SpliceAPP, to provide search and prediction of splicing errors arising from intronic mutations. This tool is based on massively parallel splicing assays on human disease-relevant intronic variants and a subsequent statistical model that learns to differentiate splicing variants (DOI: 10.1038/s41594-022-00844-1). The model is highly sensitive and accurately annotates the splicing defects of near-exon intronic variants, outperforming state-of-the-art predictive tools. SpliceAPP represents a pioneering approach to screening pathogenic intronic variants, contributing to the development of precision medicine. It also facilitates the annotation of splicing motifs. SpliceAPP is freely accessible using the link https://bc.imb.sinica.edu.tw/SpliceAPP